

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146806.1 - phase: 0
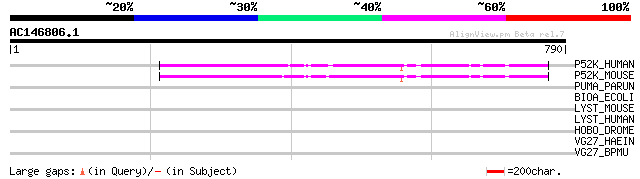
(790 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
P52K_HUMAN (O43422) 52 kDa repressor of the inhibitor of the pro... 94 1e-18
P52K_MOUSE (Q9CUX1) 52 kDa repressor of the inhibitor of the pro... 91 9e-18
PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated p... 35 0.77
BIOA_ECOLI (P12995) Adenosylmethionine-8-amino-7-oxononanoate am... 33 2.9
LYST_MOUSE (P97412) Lysosomal trafficking regulator (Beige prote... 33 3.8
LYST_HUMAN (Q99698) Lysosomal trafficking regulator (Beige homolog) 33 3.8
HOBO_DROME (P12258) Transposable element Hobo transposase 32 5.0
VG27_HAEIN (P44223) Mu-like prophage FluMu protein gp27 32 6.5
VG27_BPMU (Q9T1W7) Protein gp27 32 8.5
>P52K_HUMAN (O43422) 52 kDa repressor of the inhibitor of the
protein kinase (p58IPK-interacting protein) (58 kDa
interferon-induced protein kinase-interacting protein)
(P52rIPK) (Death associated protein 4) (THAP domain
protein 0)
Length = 761
Score = 94.4 bits (233), Expect = 1e-18
Identities = 123/570 (21%), Positives = 236/570 (40%), Gaps = 44/570 (7%)
Query: 214 SDEADDSLYKGPFLELLDTLKENNSD--VATILDSAPGNSLMTCPKIQKDLASACACEIT 271
+DE + L+ + L + N+ + + ++ N+L Q+ + C I
Sbjct: 188 ADEIPEGLFTPDNFQALLECRINSGEEVLRKRFETTAVNTLFCSKTQQRQMLEICESCIR 247
Query: 272 REIVCDIADD-VFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSL 330
E + ++ D F ++ D+ D++G E + V++R+VD ++E F+G E A+ L
Sbjct: 248 EETLREVRDSHFFSIITDDVVDIAGEEHLPVLVRFVDESHNLREEFIGFLPY-EADAEIL 306
Query: 331 KDALETMLSIN-GLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQL 389
TM++ GL+ RGQ Y +S + + + + + P A Y C + L +
Sbjct: 307 AVKFHTMITEKWGLNMEYCRGQAYIVSSGFSSKMKVVASRLLEKYPQAIYTLCSSCALNM 366
Query: 390 ALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLN 449
L VS G + + +F S Q +L +L ++ + E G L
Sbjct: 367 WLAKSVPV-MGVSVALGTIEEVCSFFHRSP--QLLL---ELDNVISVLFQNSKERGKELK 420
Query: 450 QDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPS--FDKF--GETVLLLDVL 505
+ ++W L+ L A+V + + +D + ++ + G +L +
Sbjct: 421 EICH------SQWTGRHDAFEILVELLQALVLCLDGINSDTNIRWNNYIAGRAFVLCSAV 474
Query: 506 QSFDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMR------NEG 559
FDFI + ++ +L T LQ + D+ A + L E+ +E
Sbjct: 475 SDFDFIVTIVVLKNVLSFTRAFGKNLQGQTSDVFFAAGSLTAVLHSLNEVMENIEVYHEF 534
Query: 560 WEELISRVVTICTKHEIDVPDMDAPYMEGK--KPRRVPPVSSVSNLHHYKNDCLFSVLDL 617
W E + TK +I + + GK + + S +++ +YK ++
Sbjct: 535 WFE---EATNLATKLDIQMK------LPGKFRRAHQGNLESQLTSESYYKETLSVPTVEH 585
Query: 618 QLQELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQL 677
+QEL F E++ + L+C+S + F+ ++ A++Y +D + L +L
Sbjct: 586 IIQELKDIFSEQHLKALKCLSLVPSVMGQLKFNTSEE-HHADMYRSDLPNPDT--LSAEL 642
Query: 678 HNYVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFS 737
H + K K +L L + K F V+ LLK+ +LPV ER +
Sbjct: 643 HCWRIKWKHRGKDIELPSTIYEALHLPDI---KFFPNVYALLKVLCILPVMKVENERYEN 699
Query: 738 AMKIVKSHLRNKMGDQWLNDRLVTFIERDV 767
K +K++LRN + DQ ++ + I D+
Sbjct: 700 GRKRLKAYLRNTLTDQRSSNLALLNINFDI 729
>P52K_MOUSE (Q9CUX1) 52 kDa repressor of the inhibitor of the
protein kinase (p58IPK-interacting protein) (58 kDa
interferon-induced protein kinase-interacting protein)
(P52rIPK) (THAP domain protein 0)
Length = 758
Score = 91.3 bits (225), Expect = 9e-18
Identities = 126/568 (22%), Positives = 237/568 (41%), Gaps = 39/568 (6%)
Query: 214 SDEADDSLYKGP-FLELLDTLKENNSDVATILDSAPGNSLMTCPKIQ-KDLASACACEIT 271
+DE + L+ F LL+ + +V A + + C K Q + + C I
Sbjct: 184 ADEVPEGLFAPDNFQALLECRINSGEEVLRKRFEATAVNTLFCSKTQQRHMLEICESCIR 243
Query: 272 REIVCDIADD-VFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFLGITSVKETSAKSL 330
E + ++ D F ++ D+ D++G E + V++R+VD ++E F+G E A+ L
Sbjct: 244 EETLREVRDSHFFSIITDDVVDIAGEEHLPVLVRFVDDAHNLREEFVGFLPY-EADAEIL 302
Query: 331 KDALETMLSIN-GLSFSSIRGQGYDGASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQL 389
T ++ GL+ RGQ Y +S + + + + + P A Y C + L
Sbjct: 303 AVKFHTTITEKWGLNMEYCRGQAYIVSSGFSSKMKVVASRLLEKYPQAVYTLCSSCALN- 361
Query: 390 ALVACAKTHKPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLN 449
A +A + VS G + + +F S Q +L +L ++ + E L
Sbjct: 362 AWLAKSVPVIGVSVALGTIEEVCSFFHRSP--QLLL---ELDSVISVLFQNSEERAKELK 416
Query: 450 QDSSIARAGDTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKF--GETVLLLDVLQS 507
+ I + T F L L+ ++ I+ + ++ + G +L +
Sbjct: 417 E---ICHSQWTGRHDAFEILVDLLQALVLCLDGIINSDTNVRWNNYIAGRAFVLCSAVTD 473
Query: 508 FDFIFMLYMMVEILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMR------NEGWE 561
FDFI + ++ +L T LQ + D+ A S + L E+ +E W
Sbjct: 474 FDFIVTIVVLKNVLSFTRAFGKNLQGQTSDVFFAASSLTAVLHSLNEVMENIEVYHEFWF 533
Query: 562 ELISRVVTICTKHEIDVPDMDAPYMEGK--KPRRVPPVSSVSNLHHYKNDCLFSVLDLQL 619
E + TK +I + + GK + ++ S +++ +YK+ ++ +
Sbjct: 534 E---EATNLATKLDIQMK------LPGKFRRAQQGNLESQLTSESYYKDTLSVPTVEHII 584
Query: 620 QELNARFDEENTELLQCVSCLSPAKSFSAFDVNKLLRMAELYPNDFVDVSEVELRRQLHN 679
QEL F E++ + L+C+S + F+ ++ A++Y +D + L +LH
Sbjct: 585 QELKDIFSEQHLKALKCLSLVPSVMGQLKFNTSEE-HHADMYRSDLPNPDT--LSAELHC 641
Query: 680 YVRNVKSDPKFAKLKGLSDLCAILVETNKCKTFALVFKLLKLALLLPVATASVERVFSAM 739
+ K K +L L + K F V+ LLK+ +LPV ER +
Sbjct: 642 WRIKWKHRGKDIELPSTIYEALHLPDI---KFFPNVYALLKVLCILPVMKVENERYENGR 698
Query: 740 KIVKSHLRNKMGDQWLNDRLVTFIERDV 767
K +K++LRN + DQ ++ + I D+
Sbjct: 699 KRLKAYLRNTLTDQRSSNLALLNINFDI 726
>PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated
protein (PUMA1)
Length = 1955
Score = 35.0 bits (79), Expect = 0.77
Identities = 30/102 (29%), Positives = 51/102 (49%), Gaps = 4/102 (3%)
Query: 535 DQDLLNAISLVNDTKKQLQEMRNEGWEELISRVVTICTKHEIDVPDMDAPYMEGK-KPRR 593
++DL +A++ V + KKQLQ+M E E L S + T + +D ++ A Y E K K R
Sbjct: 787 ERDLKDAMTEVEELKKQLQKMDEENSERLESVLRTKISSDTVDTSEI-AEYTEVKVKELR 845
Query: 594 VPPVSSVSNLHHYKNDC--LFSVLDLQLQELNARFDEENTEL 633
+ + L K+D +L+ +L E + + TE+
Sbjct: 846 EKYKADLERLQSNKDDLERRVQILEDELAERQRIVERQRTEM 887
>BIOA_ECOLI (P12995) Adenosylmethionine-8-amino-7-oxononanoate
aminotransferase (EC 2.6.1.62) (7,8-diamino-pelargonic
acid aminotransferase) (DAPA aminotransferase)
Length = 429
Score = 33.1 bits (74), Expect = 2.9
Identities = 17/56 (30%), Positives = 29/56 (51%)
Query: 262 LASACACEITREIVCDIADDVFCVLIDESGDVSGREQMAVVLRYVDGDGLVKERFL 317
+ A A E+ R++V + CV + +SG V+ M + L+Y G ++RFL
Sbjct: 83 ITHAPAIELCRKLVAMTPQPLECVFLADSGSVAVEVAMKMALQYWQAKGEARQRFL 138
>LYST_MOUSE (P97412) Lysosomal trafficking regulator (Beige protein)
(CHS1 homolog)
Length = 3788
Score = 32.7 bits (73), Expect = 3.8
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 235 ENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIA 279
E + D+AT+ DSA G S + + DL C REI+C +A
Sbjct: 3655 ETSGDIATVCDSAGGGSDLRLWTVNGDLVGHVHC---REIICSVA 3696
>LYST_HUMAN (Q99698) Lysosomal trafficking regulator (Beige homolog)
Length = 3801
Score = 32.7 bits (73), Expect = 3.8
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 235 ENNSDVATILDSAPGNSLMTCPKIQKDLASACACEITREIVCDIA 279
E + D+AT+ DSA G S + + DL C REI+C +A
Sbjct: 3668 ETSGDIATVCDSAGGGSDLRLWTVNGDLVGHVHC---REIICSVA 3709
>HOBO_DROME (P12258) Transposable element Hobo transposase
Length = 644
Score = 32.3 bits (72), Expect = 5.0
Identities = 43/217 (19%), Positives = 82/217 (36%), Gaps = 45/217 (20%)
Query: 309 DGLVKERFLGITSVKETSAK------SLKD-------ALETMLSINGLSFSSIRGQGYDG 355
D V+ FLGIT E K LK A ++ I GL FS + D
Sbjct: 232 DQYVQRNFLGITFHYEKEFKLCDMILGLKSMNFQKSTAENILMKIKGL-FSEFNVENIDN 290
Query: 356 ASNMRGRFGGLKTLIQNENPSAHYVHCFAHQLQLALVACAKTHKPVSGFFGKVNMLVNFI 415
+ R +K ++ ++C +H L L F + N L +
Sbjct: 291 VKFVTDRGANIKKALEGNTR----LNCSSHLLSNVLEKS----------FNEANELKKIV 336
Query: 416 RASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRWGSHFRTLTSLMTL 475
++ K + + L ++++ A TRW S+++ + S++
Sbjct: 337 KSCKKIVKYCKKSNLQH----------------TLETTLKSACPTRWNSNYKMMKSILDN 380
Query: 476 YGAIVEVIVEVGNDPSFDKFGETVLLLDVLQSFDFIF 512
+ ++ +++ E F+K +++D+L F+ IF
Sbjct: 381 WRSVDKILGEADIHVDFNK-SSLKVVVDILGDFERIF 416
>VG27_HAEIN (P44223) Mu-like prophage FluMu protein gp27
Length = 189
Score = 32.0 bits (71), Expect = 6.5
Identities = 38/160 (23%), Positives = 69/160 (42%), Gaps = 16/160 (10%)
Query: 399 KPVSGFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAG 458
K G KV++L I+++ MLRDKQ +Q L E I SGL++ +++ G
Sbjct: 4 KTTRGRASKVDLLPPNIKSTLTM--MLRDKQYSQAEILEEINNIIADSGLDESMQLSKTG 61
Query: 459 DTRWGSHFRTLTSLMTLYGAIVEVIVEVGNDPSFDKFGETVLLLDVLQSFDFIFMLYMMV 518
R+ S + + EV + + G+ LL++ +++ F
Sbjct: 62 LNRFASKMERFGKKIREAREVAEVWTKQLVEAPQSDIGK--LLMEAVKTMAF-------- 111
Query: 519 EILGITNDLSLALQRRDQDLLNAISLVNDTKKQLQEMRNE 558
L + D ++A D LN ++L+ + +Q Q + E
Sbjct: 112 -DLTLNADEAVA---NDPKFLNQLALIANRIEQAQSISEE 147
>VG27_BPMU (Q9T1W7) Protein gp27
Length = 191
Score = 31.6 bits (70), Expect = 8.5
Identities = 31/114 (27%), Positives = 56/114 (48%), Gaps = 8/114 (7%)
Query: 403 GFFGKVNMLVNFIRASNKRQEMLRDKQLAQFAKLIEEGEIETGSGLNQDSSIARAGDTRW 462
G KV++L +R + EMLRDK + Q L E + +GL + ++R+G R+
Sbjct: 7 GRASKVDLLPENVRKT--LHEMLRDKAIPQARILEEINALIEDAGLPDEMKLSRSGLNRY 64
Query: 463 GSHFRTL-TSLMTLYGAIVEVIVEVGNDPSFDKFGETV-LLLDVLQSFDFIFML 514
++ + +L + + E+G+ P GET L+L++ +S F M+
Sbjct: 65 ATNVEQVGHNLRQMREMTSALTAELGDKP----MGETTKLILEMARSQLFKAMM 114
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,720,648
Number of Sequences: 164201
Number of extensions: 3768392
Number of successful extensions: 9385
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 9375
Number of HSP's gapped (non-prelim): 10
length of query: 790
length of database: 59,974,054
effective HSP length: 118
effective length of query: 672
effective length of database: 40,598,336
effective search space: 27282081792
effective search space used: 27282081792
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146806.1


