

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146793.2 - phase: 0
(438 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
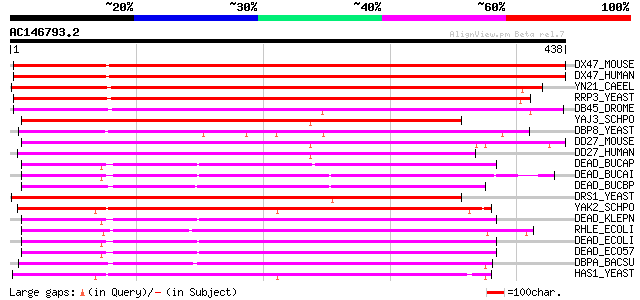
Score E
Sequences producing significant alignments: (bits) Value
DX47_MOUSE (Q9CWX9) DEAD-box protein 47 (EC 3.6.1.-) 549 e-156
DX47_HUMAN (Q9H0S4) DEAD-box protein 47 (EC 3.6.1.-) 545 e-155
YN21_CAEEL (P34580) Putative ATP-dependent RNA helicase T26G10.1... 498 e-140
RRP3_YEAST (P38712) ATP-dependent rRNA helicase RRP3 409 e-113
DB45_DROME (Q07886) Probable ATP-dependent RNA helicase Dbp45A (... 292 1e-78
YAJ3_SCHPO (Q09903) Putative ATP-dependent RNA helicase C30D11.03 270 4e-72
DBP8_YEAST (P38719) Probable ATP-dependent RNA helicase DBP8 269 9e-72
DD27_MOUSE (Q921N6) Probable ATP-dependent RNA helicase DDX27 (D... 266 1e-70
DD27_HUMAN (Q96GQ7) Probable ATP-dependent RNA helicase DDX27 (D... 266 1e-70
DEAD_BUCAP (Q8K9H6) Cold-shock DEAD-box protein A homolog (ATP-d... 258 2e-68
DEAD_BUCAI (P57453) Cold-shock DEAD-box protein A homolog (ATP-d... 258 3e-68
DEAD_BUCBP (Q89AF9) Cold-shock DEAD-box protein A homolog (ATP-d... 253 5e-67
DRS1_YEAST (P32892) Probable ATP-dependent RNA helicase DRS1 252 1e-66
YAK2_SCHPO (Q09916) Putative ATP-dependent RNA helicase C1F7.02c 250 4e-66
DEAD_KLEPN (P33906) Cold-shock DEAD-box protein A (ATP-dependent... 250 4e-66
RHLE_ECOLI (P25888) Putative ATP-dependent RNA helicase rhlE 250 6e-66
DEAD_ECOLI (P23304) Cold-shock DEAD-box protein A (ATP-dependent... 249 1e-65
DEAD_ECO57 (Q8XA87) Cold-shock DEAD-box protein A (ATP-dependent... 249 1e-65
DBPA_BACSU (P42305) ATP-dependent RNA helicase dbpA 248 2e-65
HAS1_YEAST (Q03532) Probable ATP-dependent RNA helicase HAS1 248 2e-65
>DX47_MOUSE (Q9CWX9) DEAD-box protein 47 (EC 3.6.1.-)
Length = 455
Score = 549 bits (1415), Expect = e-156
Identities = 270/435 (62%), Positives = 353/435 (81%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWAKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+CAV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQCAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHI++ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIVIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYLFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P +EEV++L ERV EA+R A +++E G KK+R D G++DD +
Sbjct: 377 RIEHLIGKKLPVFPTQDEEVMMLTERVNEAQRFARMELREHGEKKKRKREDAGDDDDKEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>DX47_HUMAN (Q9H0S4) DEAD-box protein 47 (EC 3.6.1.-)
Length = 455
Score = 545 bits (1404), Expect = e-155
Identities = 269/435 (61%), Positives = 351/435 (79%), Gaps = 2/435 (0%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E +E K+FKDLG+ + L EAC+++GW P +IQIEAIP AL+G+D+IGLA+TGSGKTGAF
Sbjct: 19 EEEETKTFKDLGVTDVLCEACDQLGWTKPTKIQIEAIPLALQGRDIIGLAETGSGKTGAF 78
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL+ALLE P+ FA V++PTRELA QISEQFEALGS IGV+ AV+VGGID + QS
Sbjct: 79 ALPILNALLETPQ--RLFALVLTPTRELAFQISEQFEALGSSIGVQSAVIVGGIDSMSQS 136
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ +AK PHII+ TPGR++DHL+NTKGF+L LKYLV+DEADR+LN DFE +++IL +IP
Sbjct: 137 LALAKKPHIIIATPGRLIDHLENTKGFNLRALKYLVMDEADRILNMDFETEVDKILKVIP 196
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
R+R+TFLFSATMT KV+KLQR L+NPVK SSKY TV+ L+Q Y F+P+K KD YLVY
Sbjct: 197 RDRKTFLFSATMTKKVQKLQRAALKNPVKCAVSSKYQTVEKLQQYYIFIPSKFKDTYLVY 256
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
IL+E+AG++ M+F TC++T+ AL+LRNLG AIP++G MSQ KRLG+LNKFK+ +I
Sbjct: 257 ILNELAGNSFMIFCSTCNNTQRTALLLRNLGFTAIPLHGQMSQSKRLGSLNKFKAKARSI 316
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
LL TDVASRGLDIP VD+V+N+DIPT+SKDYIHRVGRTARAGRSG AI+ V QY++E +
Sbjct: 317 LLATDVASRGLDIPHVDVVVNFDIPTHSKDYIHRVGRTARAGRSGKAITFVTQYDVELFQ 376
Query: 364 QIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDK 423
+IE LIGKKLP +P ++EV++L ERV EA+R A +++E G KK+R D G+ DD +
Sbjct: 377 RIEHLIGKKLPGFPTQDDEVMMLTERVAEAQRFARMELREHGEKKKRSREDAGDNDDTEG 436
Query: 424 YFGLKDRKSSKKFRR 438
G++++ + K ++
Sbjct: 437 AIGVRNKVAGGKMKK 451
>YN21_CAEEL (P34580) Putative ATP-dependent RNA helicase T26G10.1 in
chromosome III
Length = 489
Score = 498 bits (1282), Expect = e-140
Identities = 247/424 (58%), Positives = 331/424 (77%), Gaps = 7/424 (1%)
Query: 2 EEENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTG 61
+EE+ + KSF +LG+ + L +AC+++GW P +IQ A+P AL+GKD+IGLA+TGSGKTG
Sbjct: 37 DEEDVKEKSFAELGVSQPLCDACQRLGWMKPSKIQQAALPHALQGKDVIGLAETGSGKTG 96
Query: 62 AFALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQ 121
AFA+P+L +LL+ P+ FF V++PTRELA QI +QFEALGS IG+ AV+VGG+DM
Sbjct: 97 AFAIPVLQSLLDHPQA--FFCLVLTPTRELAFQIGQQFEALGSGIGLIAAVIVGGVDMAA 154
Query: 122 QSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGM 181
Q++ +A+ PHIIV TPGR++DHL+NTKGF+L LK+L++DEADR+LN DFE L++IL +
Sbjct: 155 QAMALARRPHIIVATPGRLVDHLENTKGFNLKALKFLIMDEADRILNMDFEVELDKILKV 214
Query: 182 IPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYL 241
IPRERRT+LFSATMT KV KL+R LR+P ++ SS+Y TVD LKQ Y F+P K+K+ YL
Sbjct: 215 IPRERRTYLFSATMTKKVSKLERASLRDPARVSVSSRYKTVDNLKQHYIFVPNKYKETYL 274
Query: 242 VYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDC 301
VY+L+E AG++++VF TC +T +A++LR LG++A+P++G MSQ KRLG+LNKFKS
Sbjct: 275 VYLLNEHAGNSAIVFCATCATTMQIAVMLRQLGMQAVPLHGQMSQEKRLGSLNKFKSKAR 334
Query: 302 NILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEW 361
IL+CTDVA+RGLDIP VDMVINYD+P+ SKDY+HRVGRTARAGRSG+AI++V QY++E
Sbjct: 335 EILVCTDVAARGLDIPHVDMVINYDMPSQSKDYVHRVGRTARAGRSGIAITVVTQYDVEA 394
Query: 362 YVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKE-----SGGKKRRGEGDIG 416
Y +IE +GKKL EY E EV++L ER EA A +MKE GKKRR D G
Sbjct: 395 YQKIEANLGKKLDEYKCVENEVMVLVERTQEATENARIEMKEMDEKKKSGKKRRQNDDFG 454
Query: 417 EEDD 420
+ ++
Sbjct: 455 DTEE 458
>RRP3_YEAST (P38712) ATP-dependent rRNA helicase RRP3
Length = 543
Score = 409 bits (1050), Expect = e-113
Identities = 211/415 (50%), Positives = 300/415 (71%), Gaps = 9/415 (2%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
E++ +SF +L L L++AC+ + + P IQ +AIPPALEG D+IGLA+TGSGKT AF
Sbjct: 118 EDESFESFSELNLVPELIQACKNLNYSKPTPIQSKAIPPALEGHDIIGLAQTGSGKTAAF 177
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
A+PIL+ L P ++AC+++PTRELA QI E F++LGS +GV+ +VGG++M+ Q+
Sbjct: 178 AIPILNRLWHDQEP--YYACILAPTRELAQQIKETFDSLGSLMGVRSTCIVGGMNMMDQA 235
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+ + PHII+ TPGR++DHL+NTKGFSL +LK+LV+DEADRLL+ +F L+ IL +IP
Sbjct: 236 RDLMRKPHIIIATPGRLMDHLENTKGFSLRKLKFLVMDEADRLLDMEFGPVLDRILKIIP 295
Query: 184 -RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLV 242
+ER T+LFSATMT+K++KLQR L NPVK S+KY TVDTL Q +P K+ YL+
Sbjct: 296 TQERTTYLFSATMTSKIDKLQRASLTNPVKCAVSNKYQTVDTLVQTLMVVPGGLKNTYLI 355
Query: 243 YILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCN 302
Y+L+E G T ++FTRT + L+ + L A ++G ++Q +R+G+L+ FK+G +
Sbjct: 356 YLLNEFIGKTMIIFTRTKANAERLSGLCNLLEFSATALHGDLNQNQRMGSLDLFKAGKRS 415
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWY 362
IL+ TDVA+RGLDIP+VD+V+NYDIP +SK YIHRVGRTARAGRSG +ISLV+QY+LE
Sbjct: 416 ILVATDVAARGLDIPSVDIVVNYDIPVDSKSYIHRVGRTARAGRSGKSISLVSQYDLELI 475
Query: 363 VQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKM------KESGGKKRRG 411
++IE+++GKKLP+ ++ +L L + V +A +M K + GK RRG
Sbjct: 476 LRIEEVLGKKLPKESVDKNIILTLRDSVDKANGEVVMEMNRRNKEKIARGKGRRG 530
>DB45_DROME (Q07886) Probable ATP-dependent RNA helicase Dbp45A
(DEAD-box protein 45A)
Length = 521
Score = 292 bits (747), Expect = 1e-78
Identities = 180/444 (40%), Positives = 260/444 (58%), Gaps = 12/444 (2%)
Query: 4 ENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAF 63
+ KE F+ LGL LV+ K+G K IQ + IP L G+D IG AKTGSGKT AF
Sbjct: 2 QRKEANPFQILGLRPWLVKQLTKLGLKGATPIQQKCIPAILAGQDCIGAAKTGSGKTFAF 61
Query: 64 ALPILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
ALPIL L E P + FA V++PT ELA QISEQF G +GV+ V+ GG D + +S
Sbjct: 62 ALPILERLSEEPVSH--FALVLTPTHELAYQISEQFLVAGQAMGVRVCVVSGGTDQMVES 119
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
K+ + PHI+V PGR+ DHL FS LKYLV+DEADR+LN DF+ESL+ I +P
Sbjct: 120 QKLMQRPHIVVAMPGRLADHLTGCDTFSFDNLKYLVVDEADRMLNGDFDESLSIIERCLP 179
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNP-VKIETSSKYSTVDTLKQQYRFLPAKHKDCYLV 242
+ R+ FSATM + +++ + + + S +TV+TL Q+Y +D L+
Sbjct: 180 KTRQNLFFSATMKDFIKESSIFPIASDCFEWSQDSDVATVETLDQRYLLCADYDRDMVLI 239
Query: 243 YIL----SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKS 298
L E + M+FT T +LL++ L+N+ + + ++G M Q +R+ AL++FKS
Sbjct: 240 EALRKYREENENANVMIFTNTKKYCQLLSMTLKNMEIDNVCLHGFMRQKERVAALSRFKS 299
Query: 299 GDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVN-QY 357
L+ TDVA+RGLDIP+V++V+N+ +P K+YIHRVGRTARAGR G++IS+
Sbjct: 300 NQIRTLIATDVAARGLDIPSVELVMNHMLPRTPKEYIHRVGRTARAGRKGMSISIFRFPR 359
Query: 358 ELEWYVQIEKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKR----RGEG 413
+LE IE+ I KL E+P ++ V + +V +R + ++ + +R R +
Sbjct: 360 DLELLAAIEEEINTKLTEHPIDQRMVERIFMQVNVTRRESEMQLDNNDFDERAQNYRRKT 419
Query: 414 DIGEEDDVDKYFGLKDRKSSKKFR 437
I E D D+ L +K K R
Sbjct: 420 WIMEGKDPDQMEALYRKKQKDKLR 443
>YAJ3_SCHPO (Q09903) Putative ATP-dependent RNA helicase C30D11.03
Length = 754
Score = 270 bits (691), Expect = 4e-72
Identities = 141/351 (40%), Positives = 219/351 (62%), Gaps = 4/351 (1%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
SF+ + L +++ +G++ P +IQ + IP AL GKD++G A TGSGKT AF +PIL
Sbjct: 260 SFQSMNLSRPILKGLSNLGFEVPTQIQDKTIPLALLGKDIVGAAVTGSGKTAAFIVPILE 319
Query: 70 ALLEAPRPNHFF-ACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAK 128
LL P+ ++ PTRELA+Q + S + + +GG+ + Q ++ K
Sbjct: 320 RLLYRPKKVPTTRVLILCPTRELAMQCHSVATKIASFTDIMVCLCIGGLSLKLQEQELRK 379
Query: 129 LPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRT 188
P I++ TPGR +DH++N++GF++ ++ +V+DEADR+L + F + LNEI+ P+ R+T
Sbjct: 380 RPDIVIATPGRFIDHMRNSQGFTVENIEIMVMDEADRMLEDGFADELNEIIQACPKSRQT 439
Query: 189 FLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQY-RFLPAKH--KDCYLVYIL 245
LFSATMT+KV+ L R+ L PV++ +K +T L Q++ R P + + L+Y+
Sbjct: 440 MLFSATMTDKVDDLIRLSLNRPVRVFVDNKKTTAKLLTQEFVRVRPQRELLRPAMLIYLC 499
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
E+ +++F R+ + +I L L A I+G +SQ +R+ AL F+ G CN LL
Sbjct: 500 KELFHRRTIIFFRSKAFAHKMRVIFGLLSLNATEIHGSLSQEQRVRALEDFRDGKCNYLL 559
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQ 356
TDVASRG+DI +++VINY+ P + Y+HRVGRTARAGRSG AI+L +
Sbjct: 560 ATDVASRGIDIKGIEVVINYEAPATHEVYLHRVGRTARAGRSGRAITLAGE 610
>DBP8_YEAST (P38719) Probable ATP-dependent RNA helicase DBP8
Length = 431
Score = 269 bits (688), Expect = 9e-72
Identities = 165/419 (39%), Positives = 249/419 (59%), Gaps = 18/419 (4%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPI 67
M FK LGL + L E+ + P IQ IP LEG+D IG AKTGSGKT AFA P+
Sbjct: 1 MADFKSLGLSKWLTESLRAMKITQPTAIQKACIPKILEGRDCIGGAKTGSGKTIAFAGPM 60
Query: 68 LHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIA 127
L E P + F V++PTRELA+QI+EQF ALGS + ++ +V+VGG +VQQ++ +
Sbjct: 61 LTKWSEDP--SGMFGVVLTPTRELAMQIAEQFTALGSSMNIRVSVIVGGESIVQQALDLQ 118
Query: 128 KLPHIIVGTPGRVLDHLKNTKGFS---LARLKYLVLDEADRLLNEDFEESLNEILGMIPR 184
+ PH I+ TPGR+ H+ ++ + L R KYLVLDEAD LL F + L + +P
Sbjct: 119 RKPHFIIATPGRLAHHIMSSGDDTVGGLMRAKYLVLDEADILLTSTFADHLATCISALPP 178
Query: 185 E--RRTFLFSATMTNKVEKLQRVCLRN------PVKIETSSKYSTVDTLKQQYRFLPAKH 236
+ R+T LF+AT+T++V+ LQ ++ ++E+ + TLK +Y +P
Sbjct: 179 KDKRQTLLFTATITDQVKSLQNAPVQKGKPPLFAYQVESVDNVAIPSTLKIEYILVPEHV 238
Query: 237 KDCYLVYILS--EMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALN 294
K+ YL +L+ E T+++F + +L L+ L ++ ++ M Q +R +L+
Sbjct: 239 KEAYLYQLLTCEEYENKTAIIFVNRTMTAEILRRTLKQLEVRVASLHSQMPQQERTNSLH 298
Query: 295 KFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLV 354
+F++ IL+ TDVASRGLDIP V++V+NYDIP++ +IHR GRTARAGR G AIS V
Sbjct: 299 RFRANAARILIATDVASRGLDIPTVELVVNYDIPSDPDVFIHRSGRTARAGRIGDAISFV 358
Query: 355 NQYELEWYVQIEKLIGKKLPEYPANEEEVLLLE--ERVGEAKRLAATKM-KESGGKKRR 410
Q ++ IE I KK+ E + ++ + +V +AKR + M KE+ G+++R
Sbjct: 359 TQRDVSRIQAIEDRINKKMTETNKVHDTAVIRKALTKVTKAKRESLMAMQKENFGERKR 417
>DD27_MOUSE (Q921N6) Probable ATP-dependent RNA helicase DDX27
(DEAD-box protein 27)
Length = 760
Score = 266 bits (679), Expect = 1e-70
Identities = 167/468 (35%), Positives = 252/468 (53%), Gaps = 39/468 (8%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
SF+D+ L L++A +G+K P IQ IP L GKD+ A TG+GKT AFALP+L
Sbjct: 185 SFQDMNLSRPLLKAITAMGFKQPTPIQKACIPVGLLGKDICACAATGTGKTAAFALPVLE 244
Query: 70 ALLEAPRPNHFF-ACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKIAK 128
L+ PR V+ PTREL IQ+ + L + + VGG+D+ Q +
Sbjct: 245 RLIYKPRQAAVTRVLVLVPTRELGIQVHSVTKQLAQFCSITTCLAVGGLDVKSQEAALRA 304
Query: 129 LPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERRT 188
P I++ TPGR++DHL N F L+ ++ L+LDEAD++L+E FEE + EI+ M R+T
Sbjct: 305 APDILIATPGRLIDHLHNCPSFHLSSIEVLILDEADKMLDEYFEEQMKEIIRMCSHHRQT 364
Query: 189 FLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQY-RFLPAKH--KDCYLVYIL 245
LFSATMT++V+ L V L+NPV+I +S L+Q++ R P + ++ + +L
Sbjct: 365 MLFSATMTDEVKDLASVSLKNPVRIFVNSNTDVAPFLRQEFIRIRPNREGDREAIVAALL 424
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
M+FT+T + ++L LGL+ ++G++SQ +RL AL +FK +IL+
Sbjct: 425 MRTFTDHVMLFTQTKKQAHRMHILLGLLGLQVGELHGNLSQTQRLEALRRFKDEQIDILV 484
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLDI V VIN+ +P K Y+HRVGRTARAGR+G ++SLV + E + +I
Sbjct: 485 ATDVAARGLDIEGVKTVINFTMPNTVKHYVHRVGRTARAGRAGRSVSLVGEEERKMLKEI 544
Query: 366 EK-----LIGKKLP----------------------EYPANEEEVLLLEERVGEAKRLAA 398
K + + LP + A E+E+ E ++ A+RL A
Sbjct: 545 VKAAKAPVKARILPQDVILKFRDKIEKLEKDVYAVLQLEAEEKEMQQSEAQIDTAQRLLA 604
Query: 399 TKMKESGGKKRRGEGDIGEEDDVDKY--------FGLKDRKSSKKFRR 438
+ + + R EE +K L+ +K KKF +
Sbjct: 605 KGKETADQEPERSWFQTKEERKKEKIAKALQEFDLALRGKKKRKKFMK 652
>DD27_HUMAN (Q96GQ7) Probable ATP-dependent RNA helicase DDX27
(DEAD-box protein 27) (HSPC259) (PP3241)
Length = 796
Score = 266 bits (679), Expect = 1e-70
Identities = 149/365 (40%), Positives = 219/365 (59%), Gaps = 4/365 (1%)
Query: 7 EMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALP 66
E SF+D+ L L++A +G+K P IQ IP L GKD+ A TG+GKT AFALP
Sbjct: 216 ENLSFQDMNLSRPLLKAITAMGFKQPTPIQKACIPVGLLGKDICACAATGTGKTAAFALP 275
Query: 67 ILHALLEAPRPNHFF-ACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVK 125
+L L+ PR V+ PTREL IQ+ L + + VGG+D+ Q
Sbjct: 276 VLERLIYKPRQAPVTRVLVLVPTRELGIQVHSVTRQLAQFCNITTCLAVGGLDVKSQEAA 335
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ P I++ TPGR++DHL N F L+ ++ L+LDEADR+L+E FEE + EI+ M
Sbjct: 336 LRAAPDILIATPGRLIDHLHNCPSFHLSSIEVLILDEADRMLDEYFEEQMKEIIRMCSHH 395
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQY-RFLPAKH--KDCYLV 242
R+T LFSATMT++V+ L V L+NPV+I +S L+Q++ R P + ++ +
Sbjct: 396 RQTMLFSATMTDEVKDLASVSLKNPVRIFVNSNTDVAPFLRQEFIRIRPNREGDREAIVA 455
Query: 243 YILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCN 302
+L+ M+FT+T + ++L +GL+ ++G++SQ +RL AL +FK +
Sbjct: 456 ALLTRTFTDHVMLFTQTKKQAHRMHILLGLMGLQVGELHGNLSQTQRLEALRRFKDEQID 515
Query: 303 ILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWY 362
IL+ TDVA+RGLDI V VIN+ +P K Y+HRVGRTARAGR+G ++SLV + E +
Sbjct: 516 ILVATDVAARGLDIEGVKTVINFTMPNTIKHYVHRVGRTARAGRAGRSVSLVGEDERKML 575
Query: 363 VQIEK 367
+I K
Sbjct: 576 KEIVK 580
>DEAD_BUCAP (Q8K9H6) Cold-shock DEAD-box protein A homolog
(ATP-dependent RNA helicase deaD homolog)
Length = 601
Score = 258 bits (659), Expect = 2e-68
Identities = 151/379 (39%), Positives = 224/379 (58%), Gaps = 11/379 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F LGL ++++ K+G+ P IQ IP LEG+D++G+A+TGSGKT AF+LP+LH
Sbjct: 7 TFSFLGLNPFIIKSLSKMGYVKPSPIQAACIPLLLEGRDVLGMAQTGSGKTAAFSLPLLH 66
Query: 70 AL---LEAPRPNHFFACVMSPTRELAIQISEQFEALGSEI-GVKCAVLVGGIDMVQQSVK 125
L L+AP+ V++PTRELA+Q++E F I G+ L GG Q
Sbjct: 67 NLNINLKAPQ-----ILVLAPTRELAVQVAEAFSDFSKYIMGIHVLPLYGGQRYEVQLRA 121
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ + P I+VGTPGR+LDHLK +L+ L LVLDEAD +L F E + I+ IP+E
Sbjct: 122 LRQGPQIVVGTPGRLLDHLKRGT-LNLSNLYALVLDEADEMLRMGFIEDVETIMSQIPKE 180
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
+T LFSATM + ++ + ++NP +I+ S +T +KQ Y + + D L+ L
Sbjct: 181 HQTALFSATMPEAIRRISKRFMKNPQEIKIQSNITTRPDIKQSYWMVYGRKTDA-LIRFL 239
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
S +++F +T ++T ++ L G + +NG M+Q R L + KSG +IL+
Sbjct: 240 EVEDFSATIIFVKTKNATLEVSEALERNGYNSAALNGDMNQALREQTLERLKSGRLDILI 299
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLD+ + VINYDIP +S+ Y+HR+GRT RAGR+G A+ V E I
Sbjct: 300 ATDVAARGLDVDRISFVINYDIPMDSESYVHRIGRTGRAGRAGRALLFVENRERRLLRNI 359
Query: 366 EKLIGKKLPEYPANEEEVL 384
E+ I + +PE + EVL
Sbjct: 360 ERTINQTIPEVQLPKIEVL 378
>DEAD_BUCAI (P57453) Cold-shock DEAD-box protein A homolog
(ATP-dependent RNA helicase deaD homolog)
Length = 601
Score = 258 bits (658), Expect = 3e-68
Identities = 156/425 (36%), Positives = 242/425 (56%), Gaps = 27/425 (6%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F LGL ++++ ++G+ P IQ IP LEG+D++G+A+TGSGKT AF+LP+LH
Sbjct: 7 TFSFLGLNPFIIQSLNEMGYVKPSPIQASCIPLLLEGRDVLGMAQTGSGKTAAFSLPLLH 66
Query: 70 AL---LEAPRPNHFFACVMSPTRELAIQISEQFEALGSE-IGVKCAVLVGGIDMVQQSVK 125
L L+AP+ V++PTRELA+Q++E F IG+ L GG Q
Sbjct: 67 NLNINLKAPQ-----ILVLAPTRELAVQVAEAFSDFSKYMIGIHVLPLYGGQRYELQLRA 121
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ + P I+VGTPGR+LDHLK +L+ L LVLDEAD +L F E + I+ IP+E
Sbjct: 122 LRQGPQIVVGTPGRLLDHLKRGT-LNLSNLHGLVLDEADEMLRMGFIEDVETIMAQIPKE 180
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
+T LFSATM + ++ + +RNP +I+ S +T +KQ Y + + D + ++
Sbjct: 181 HQTALFSATMPEAIRRISKRFMRNPKEIKIQSNITTRPDIKQSYWMVYGRKTDALIRFLE 240
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+E +T ++F RT ++T ++ L G + +NG M+Q R L + K+G +IL+
Sbjct: 241 AEDFSAT-IIFVRTKNATLEVSEALERNGYNSAALNGDMNQALREQTLERLKNGRLDILI 299
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLD+ + VINYDIP +S+ Y+HR+GRT RAGR+G A+ V E I
Sbjct: 300 ATDVAARGLDVDRISFVINYDIPMDSESYVHRIGRTGRAGRAGRALLFVENRERRLLRNI 359
Query: 366 EKLIGKKLPEYPANEEEVLLLEERVGEAKRLAATKMKESGGKKRRGEGDIGEEDDVDKYF 425
E+ + + +PE + E LL + R+ + + ++ E D+D+Y
Sbjct: 360 ERTMKQSIPEVQLPKVE-LLCQRRLEQFAKKVQQQL---------------ESRDLDEYS 403
Query: 426 GLKDR 430
L D+
Sbjct: 404 ALLDK 408
>DEAD_BUCBP (Q89AF9) Cold-shock DEAD-box protein A homolog
(ATP-dependent RNA helicase deaD homolog)
Length = 602
Score = 253 bits (647), Expect = 5e-67
Identities = 147/368 (39%), Positives = 213/368 (56%), Gaps = 7/368 (1%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F GL ++ A +G+ P IQ IP ++GKD++G+A+TGSGKT AFALP+LH
Sbjct: 7 TFSSFGLNSCIITALNDIGYVQPSPIQAACIPYLIKGKDVLGMAQTGSGKTAAFALPLLH 66
Query: 70 AL-LEAPRPNHFFACVMSPTRELAIQISEQFEALGSE-IGVKCAVLVGGIDMVQQSVKIA 127
+ L+ P V++PTRELA+Q++E F + IGV L GG Q +
Sbjct: 67 NIKLDVRVPQ---ILVLTPTRELAVQVAEAFSNFSKKLIGVHVLALYGGQRYDLQLKSLR 123
Query: 128 KLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRERR 187
K P IIVGTPGR+LDHLK + SL+ L LVLDEAD +L F E + I+ IP +
Sbjct: 124 KGPQIIVGTPGRLLDHLKR-RTLSLSNLHSLVLDEADEMLRMGFIEDVETIMTEIPDRHQ 182
Query: 188 TFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYILSE 247
T LFSATM + ++ R ++NP +I S +T ++Q Y + K D + ++ +E
Sbjct: 183 TALFSATMPEAIRRISRRFMKNPKEIRIQSNITTRPDIQQSYWMVYGKKTDALIRFLEAE 242
Query: 248 MAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILLCT 307
+T ++F RT ++T ++ +L G + +NG M+Q R L K K G +IL+ T
Sbjct: 243 DFSAT-IIFVRTKNATLEVSEVLERYGYNSAALNGDMNQSLREQTLEKLKDGRLDILIAT 301
Query: 308 DVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQIEK 367
DVA+RGLD+ + VINYDIP +S+ Y+HR+GRT RAGR G A+ V E IE+
Sbjct: 302 DVAARGLDVDRISFVINYDIPMDSESYVHRIGRTGRAGRKGKALLFVENRERRLLRNIER 361
Query: 368 LIGKKLPE 375
+ + E
Sbjct: 362 AMNISISE 369
>DRS1_YEAST (P32892) Probable ATP-dependent RNA helicase DRS1
Length = 752
Score = 252 bits (644), Expect = 1e-66
Identities = 139/363 (38%), Positives = 222/363 (60%), Gaps = 8/363 (2%)
Query: 2 EEENKEM-KSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKT 60
+E K+M ++F L L +++ +G+ P IQ IP AL GKD+I A TGSGKT
Sbjct: 223 DEAKKQMYENFNSLSLSRPVLKGLASLGYVKPSPIQSATIPIALLGKDIIAGAVTGSGKT 282
Query: 61 GAFALPILHALLEAP-RPNHFFACVMSPTRELAIQISEQFEALGSEI-GVKCAVLVGGID 118
AF +PI+ LL P + V+ PTRELAIQ+++ + + + G+ + VGG++
Sbjct: 283 AAFMIPIIERLLYKPAKIASTRVIVLLPTRELAIQVADVGKQIARFVSGITFGLAVGGLN 342
Query: 119 MVQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEI 178
+ QQ + P I++ TPGR +DH++N+ F++ ++ LV+DEADR+L E F++ LNEI
Sbjct: 343 LRQQEQMLKSRPDIVIATPGRFIDHIRNSASFNVDSVEILVMDEADRMLEEGFQDELNEI 402
Query: 179 LGMIPRERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQY-RFLPAKHK 237
+G++P R+ LFSATM +K++ L + L+ PV+I L Q++ R H
Sbjct: 403 MGLLPSNRQNLLFSATMNSKIKSLVSLSLKKPVRIMIDPPKKAATKLTQEFVRIRKRDHL 462
Query: 238 DCYLVYILSEMAGSTS----MVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGAL 293
L++ L T +VF ++ L +I+ LG+ ++G ++Q +RL ++
Sbjct: 463 KPALLFNLIRKLDPTGQKRIVVFVARKETAHRLRIIMGLLGMSVGELHGSLTQEQRLDSV 522
Query: 294 NKFKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISL 353
NKFK+ + +L+CTD+ASRGLDIP +++VINYD+P + + Y+HRVGRTARAGR G +++
Sbjct: 523 NKFKNLEVPVLICTDLASRGLDIPKIEVVINYDMPKSYEIYLHRVGRTARAGREGRSVTF 582
Query: 354 VNQ 356
V +
Sbjct: 583 VGE 585
>YAK2_SCHPO (Q09916) Putative ATP-dependent RNA helicase C1F7.02c
Length = 578
Score = 250 bits (639), Expect = 4e-66
Identities = 150/384 (39%), Positives = 235/384 (61%), Gaps = 12/384 (3%)
Query: 7 EMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALP 66
+++ F DL L E++ +A +++G++T EIQ +IPP L G+D++G AKTGSGKT AF +P
Sbjct: 87 DIEKFSDLQLSENIQKAIKEMGFETMTEIQKRSIPPLLAGRDVLGAAKTGSGKTLAFLIP 146
Query: 67 ---ILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
+L+AL PR N ++SPTRELA+QI + L +++GG + ++
Sbjct: 147 TIEMLYALKFKPR-NGTGVIIISPTRELALQIFGVAKELLKYHHQTFGIVIGGANRRAEA 205
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
K+ K +++V TPGR+LDHL+NTKGF L+ LV+DEADR+L FE+ + +I+ ++P
Sbjct: 206 DKLVKGVNLLVATPGRLLDHLQNTKGFVFRNLRSLVIDEADRILEIGFEDEMRQIMKILP 265
Query: 184 RE-RRTFLFSATMTNKVEKLQRVCLRNP---VKIETSSKYSTVDTLKQQYRFLPAKHKDC 239
E R+T LFSAT T KVE L R+ L+ V +++ STV+ L+Q Y + + +
Sbjct: 266 SENRQTLLFSATQTTKVEDLARISLKPGPLYVNVDSGKPTSTVEGLEQGYVVVDSDKRFL 325
Query: 240 YLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSG 299
L L +VF +C S + +A +L + L + ++G Q +R +F +
Sbjct: 326 LLFSFLKRNLKKKVIVFMSSCASVKYMAELLNYIDLPVLDLHGKQKQQRRTNTFFEFCNA 385
Query: 300 DCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGR-SGVAISLVNQYE 358
+ ILLCT+VA+RGLDIPAVD ++ YD P + +DYIHRVGRTAR + +G ++ + E
Sbjct: 386 EKGILLCTNVAARGLDIPAVDWIVQYDPPDDPRDYIHRVGRTARGTKGTGKSLMFLAPSE 445
Query: 359 LEW--YVQIEKLIGKKLPEYPANE 380
L + Y++ K+ + E+PAN+
Sbjct: 446 LGFLRYLKTAKVSLNEF-EFPANK 468
>DEAD_KLEPN (P33906) Cold-shock DEAD-box protein A (ATP-dependent
RNA helicase deaD)
Length = 642
Score = 250 bits (639), Expect = 4e-66
Identities = 146/379 (38%), Positives = 216/379 (56%), Gaps = 10/379 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F DLGL ++EA +G++ P IQ E IP L+G+D++G+A+TGSGKT AF+LP+L+
Sbjct: 6 TFADLGLKAPILEALTDLGYEKPSPIQAECIPHLLDGRDVLGMAQTGSGKTAAFSLPLLN 65
Query: 70 AL---LEAPRPNHFFACVMSPTRELAIQISEQFEALGSEI-GVKCAVLVGGIDMVQQSVK 125
+ L AP+ V++PTRELA+Q++E + GV L GG Q
Sbjct: 66 NIDPELRAPQ-----ILVLAPTRELAVQVAEAMTEFSKHMRGVNVVALYGGQRYDVQLRA 120
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ + P I+VGTPGR+LDHLK L++L LVLDEAD +L F E + I+ IP
Sbjct: 121 LRQGPQIVVGTPGRLLDHLKRGT-LDLSKLSGLVLDEADEMLRMGFIEDVETIMAQIPEG 179
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
+T LFSATM + ++ R ++ P ++ S +T + Q Y K+ LV L
Sbjct: 180 HQTALFSATMPEAIRRITRRFMKEPQEVRIQSSVTTRPDISQSYWTAYGMRKNEALVRFL 239
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+++F RT ++T +A L G + +NG M+Q R AL + K G +IL+
Sbjct: 240 EAEDFDAAIIFVRTKNATLEVAEALERNGYNSAALNGDMNQALREQALERLKDGRLDILI 299
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLD+ + +V+NYDIP +S+ Y+HR+GRT RAGR+G A+ V E I
Sbjct: 300 ATDVAARGLDVERISLVVNYDIPMDSESYVHRIGRTGRAGRAGRALLFVENRERRLLRNI 359
Query: 366 EKLIGKKLPEYPANEEEVL 384
E+ + +PE E+L
Sbjct: 360 ERTMKLTIPEVELPNAELL 378
>RHLE_ECOLI (P25888) Putative ATP-dependent RNA helicase rhlE
Length = 454
Score = 250 bits (638), Expect = 6e-66
Identities = 150/417 (35%), Positives = 234/417 (55%), Gaps = 16/417 (3%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
SF LGL ++ A + G++ P IQ +AIP LEG+DL+ A+TG+GKT F LP+L
Sbjct: 2 SFDSLGLSPDILRAVAEQGYREPTPIQQQAIPAVLEGRDLMASAQTGTGKTAGFTLPLLQ 61
Query: 70 ALL------EAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQS 123
L+ + RP A +++PTRELA QI E + ++ V+ GG+ + Q
Sbjct: 62 HLITRQPHAKGRRPVR--ALILTPTRELAAQIGENVRDYSKYLNIRSLVVFGGVSINPQM 119
Query: 124 VKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIP 183
+K+ ++V TPGR+LD L++ L +++ LVLDEADR+L+ F + +L +P
Sbjct: 120 MKLRGGVDVLVATPGRLLD-LEHQNAVKLDQVEILVLDEADRMLDMGFIHDIRRVLTKLP 178
Query: 184 RERRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVY 243
+R+ LFSAT ++ ++ L L NP++IE + + + D + Q F+ K K L +
Sbjct: 179 AKRQNLLFSATFSDDIKALAEKLLHNPLEIEVARRNTASDQVTQHVHFVDKKRKRELLSH 238
Query: 244 ILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNI 303
++ + +VFTRT LA L G+++ I+G+ SQ R AL FKSGD +
Sbjct: 239 MIGKGNWQQVLVFTRTKHGANHLAEQLNKDGIRSAAIHGNKSQGARTRALADFKSGDIRV 298
Query: 304 LLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYV 363
L+ TD+A+RGLDI + V+NY++P +DY+HR+GRT RA +G A+SLV E +
Sbjct: 299 LVATDIAARGLDIEELPHVVNYELPNVPEDYVHRIGRTGRAAATGEALSLVCVDEHKLLR 358
Query: 364 QIEKLIGKKLPE--YPANEEEVLLLEERVGEAKRLAATKMKESGG-----KKRRGEG 413
IEKL+ K++P P E + + E + ++ + GG + RRGEG
Sbjct: 359 DIEKLLKKEIPRIAIPGYEPDPSIKAEPIQNGRQQRGGGGRGQGGGRGQQQPRRGEG 415
>DEAD_ECOLI (P23304) Cold-shock DEAD-box protein A (ATP-dependent
RNA helicase deaD)
Length = 628
Score = 249 bits (636), Expect = 1e-65
Identities = 146/379 (38%), Positives = 215/379 (56%), Gaps = 10/379 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F DLGL ++EA +G++ P IQ E IP L G+D++G+A+TGSGKT AF+LP+L
Sbjct: 6 TFADLGLKAPILEALNDLGYEKPSPIQAECIPHLLNGRDVLGMAQTGSGKTAAFSLPLLQ 65
Query: 70 AL---LEAPRPNHFFACVMSPTRELAIQISEQFEALGSEI-GVKCAVLVGGIDMVQQSVK 125
L L+AP+ V++PTRELA+Q++E + GV L GG Q
Sbjct: 66 NLDPELKAPQ-----ILVLAPTRELAVQVAEAMTDFSKHMRGVNVVALYGGQRYDVQLRA 120
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ + P I+VGTPGR+LDHLK L++L LVLDEAD +L F E + I+ IP
Sbjct: 121 LRQGPQIVVGTPGRLLDHLKRGT-LDLSKLSGLVLDEADEMLRMGFIEDVETIMAQIPEG 179
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
+T LFSATM + ++ R ++ P ++ S +T + Q Y + K+ LV L
Sbjct: 180 HQTALFSATMPEAIRRITRRFMKEPQEVRIQSSVTTRPDISQSYWTVWGMRKNEALVRFL 239
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+++F RT ++T +A L G + +NG M+Q R L + K G +IL+
Sbjct: 240 EAEDFDAAIIFVRTKNATLEVAEALERNGYNSAALNGDMNQALREQTLERLKDGRLDILI 299
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLD+ + +V+NYDIP +S+ Y+HR+GRT RAGR+G A+ V E I
Sbjct: 300 ATDVAARGLDVERISLVVNYDIPMDSESYVHRIGRTGRAGRAGRALLFVENRERRLLRNI 359
Query: 366 EKLIGKKLPEYPANEEEVL 384
E+ + +PE E+L
Sbjct: 360 ERTMKLTIPEVELPNAELL 378
>DEAD_ECO57 (Q8XA87) Cold-shock DEAD-box protein A (ATP-dependent
RNA helicase deaD)
Length = 628
Score = 249 bits (636), Expect = 1e-65
Identities = 146/379 (38%), Positives = 215/379 (56%), Gaps = 10/379 (2%)
Query: 10 SFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPILH 69
+F DLGL ++EA +G++ P IQ E IP L G+D++G+A+TGSGKT AF+LP+L
Sbjct: 6 TFADLGLKAPILEALNDLGYEKPSPIQAECIPHLLNGRDVLGMAQTGSGKTAAFSLPLLQ 65
Query: 70 AL---LEAPRPNHFFACVMSPTRELAIQISEQFEALGSEI-GVKCAVLVGGIDMVQQSVK 125
L L+AP+ V++PTRELA+Q++E + GV L GG Q
Sbjct: 66 NLDPELKAPQ-----ILVLAPTRELAVQVAEAMTDFSKHMRGVNVVALYGGQRYDVQLRA 120
Query: 126 IAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ + P I+VGTPGR+LDHLK L++L LVLDEAD +L F E + I+ IP
Sbjct: 121 LRQGPQIVVGTPGRLLDHLKRGT-LDLSKLSGLVLDEADEMLRMGFIEDVETIMAQIPEG 179
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
+T LFSATM + ++ R ++ P ++ S +T + Q Y + K+ LV L
Sbjct: 180 HQTALFSATMPEAIRRITRRFMKEPQEVRIQSSVTTRPDISQSYWTVWGMRKNEALVRFL 239
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+++F RT ++T +A L G + +NG M+Q R L + K G +IL+
Sbjct: 240 EAEDFDAAIIFVRTKNATLEVAEALERNGYNSAALNGDMNQALREQTLERLKDGRLDILI 299
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RGLD+ + +V+NYDIP +S+ Y+HR+GRT RAGR+G A+ V E I
Sbjct: 300 ATDVAARGLDVERISLVVNYDIPMDSESYVHRIGRTGRAGRAGRALLFVENRERRLLRNI 359
Query: 366 EKLIGKKLPEYPANEEEVL 384
E+ + +PE E+L
Sbjct: 360 ERTMKLTIPEVELPNAELL 378
>DBPA_BACSU (P42305) ATP-dependent RNA helicase dbpA
Length = 479
Score = 248 bits (634), Expect = 2e-65
Identities = 144/378 (38%), Positives = 214/378 (56%), Gaps = 9/378 (2%)
Query: 8 MKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGAFALPI 67
M FK+ + ++ A E +G+ P ++Q IP ALE KDL+ ++TGSGKT +F +P+
Sbjct: 1 MSHFKNYQISHDILRALEGLGYTEPTKVQQSVIPAALERKDLVVKSQTGSGKTASFGIPL 60
Query: 68 LH-ALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDMVQQSVKI 126
A + +P A +++PTRELA+Q+ E +G +K + G +Q ++
Sbjct: 61 CELANWDENKPQ---ALILTPTRELAVQVKEDITNIGRFKRIKATAVFGKSSFDKQKAEL 117
Query: 127 AKLPHIIVGTPGRVLDHLKNTKG-FSLARLKYLVLDEADRLLNEDFEESLNEILGMIPRE 185
+ HI+VGTPGRVLDH++ KG L RL YLV+DEAD +LN F E + I+ +P E
Sbjct: 118 KQKSHIVVGTPGRVLDHIE--KGTLPLDRLSYLVIDEADEMLNMGFIEQVEAIIKHLPTE 175
Query: 186 RRTFLFSATMTNKVEKLQRVCLRNPVKIETSSKYSTVDTLKQQYRFLPAKHKDCYLVYIL 245
R T LFSAT+ +EKL R ++NP IE + T ++ + ++K L +L
Sbjct: 176 RTTMLFSATLPQDIEKLSRQYMQNPEHIEVKAAGLTTRNIEHAVIQVREENKFSLLKDVL 235
Query: 246 SEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNKFKSGDCNILL 305
+ ++F RT + L L +LG I+G M Q R +N+FK G+ L+
Sbjct: 236 MTENPDSCIIFCRTKEHVNQLTDELDDLGYPCDKIHGGMIQEDRFDVMNEFKRGEYRYLV 295
Query: 306 CTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGRSGVAISLVNQYELEWYVQI 365
TDVA+RG+DI + +VINYD+P + Y+HR GRT RAG G AIS V +E + I
Sbjct: 296 ATDVAARGIDIENISLVINYDLPLEKESYVHRTGRTGRAGNKGKAISFVTAFEKRFLADI 355
Query: 366 EKLIGKKLP--EYPANEE 381
E+ IG ++P E P+ EE
Sbjct: 356 EEYIGFEIPKIEAPSQEE 373
>HAS1_YEAST (Q03532) Probable ATP-dependent RNA helicase HAS1
Length = 505
Score = 248 bits (633), Expect = 2e-65
Identities = 149/390 (38%), Positives = 227/390 (58%), Gaps = 16/390 (4%)
Query: 3 EENKEMKSFKDLGLPESLVEACEKVGWKTPLEIQIEAIPPALEGKDLIGLAKTGSGKTGA 62
E+ ++ F++L L + ++A EK+G+ T +Q IPP L G+D++G AKTGSGKT A
Sbjct: 36 EQTTCVEKFEELKLSQPTLKAIEKMGFTTMTSVQARTIPPLLAGRDVLGAAKTGSGKTLA 95
Query: 63 FALP---ILHALLEAPRPNHFFACVMSPTRELAIQISEQFEALGSEIGVKCAVLVGGIDM 119
F +P +LH+L PR N V++PTRELA+QI L +++GG +
Sbjct: 96 FLIPAIELLHSLKFKPR-NGTGIIVITPTRELALQIFGVARELMEFHSQTFGIVIGGANR 154
Query: 120 VQQSVKIAKLPHIIVGTPGRVLDHLKNTKGFSLARLKYLVLDEADRLLNEDFEESLNEIL 179
Q++ K+ K ++++ TPGR+LDHL+NTKGF LK L++DEADR+L FE+ + +I+
Sbjct: 155 RQEAEKLMKGVNMLIATPGRLLDHLQNTKGFVFKNLKALIIDEADRILEIGFEDEMRQII 214
Query: 180 GMIPRE-RRTFLFSATMTNKVEKLQRVCLRNP---VKIETSSKYSTVDTLKQQYRFLPAK 235
++P E R++ LFSAT T KVE L R+ LR + + + ST D L+Q Y +
Sbjct: 215 KILPNEDRQSMLFSATQTTKVEDLARISLRPGPLFINVVPETDNSTADGLEQGYVVCDSD 274
Query: 236 HKDCYLVYILSEMAGSTSMVFTRTCDSTRLLALILRNLGLKAIPINGHMSQPKRLGALNK 295
+ L L +VF +C+S + A +L + L + ++G Q KR +
Sbjct: 275 KRFLLLFSFLKRNQKKKIIVFLSSCNSVKYYAELLNYIDLPVLELHGKQKQQKRTNTFFE 334
Query: 296 FKSGDCNILLCTDVASRGLDIPAVDMVINYDIPTNSKDYIHRVGRTARAGR-SGVAISLV 354
F + + IL+CTDVA+RGLDIPAVD +I +D P + +DYIHRVGRTAR + G ++ +
Sbjct: 335 FCNAERGILICTDVAARGLDIPAVDWIIQFDPPDDPRDYIHRVGRTARGTKGKGKSLMFL 394
Query: 355 NQYELEWYVQIEKLIGKKLP----EYPANE 380
EL + + L K+P E+P N+
Sbjct: 395 TPNELGF---LRYLKASKVPLNEYEFPENK 421
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.137 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,765,079
Number of Sequences: 164201
Number of extensions: 2202768
Number of successful extensions: 6984
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 250
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 6016
Number of HSP's gapped (non-prelim): 418
length of query: 438
length of database: 59,974,054
effective HSP length: 113
effective length of query: 325
effective length of database: 41,419,341
effective search space: 13461285825
effective search space used: 13461285825
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146793.2


