

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.8 + phase: 0
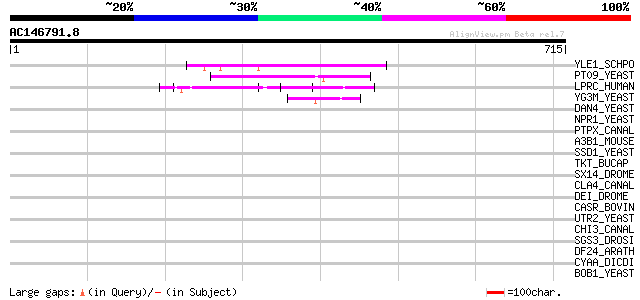
(715 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I 55 6e-07
PT09_YEAST (P32522) PET309 protein, mitochondrial precursor 53 2e-06
LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP13... 49 5e-05
YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR... 47 2e-04
DAN4_YEAST (P47179) Cell wall protein DAN4 precursor 42 0.007
NPR1_YEAST (P22211) Nitrogen permease reactivator protein (EC 2.... 37 0.18
PTPX_CANAL (P43078) Probable protein-tyrosine phosphatase (EC 3.... 37 0.24
A3B1_MOUSE (Q9Z1T1) Adapter-related protein complex 3 beta 1 sub... 36 0.40
SSD1_YEAST (P24276) SSD1 protein (SRK1 protein) 35 0.53
TKT_BUCAP (Q8KA26) Transketolase (EC 2.2.1.1) (TK) 35 0.90
SX14_DROME (P40656) Putative transcription factor SOX-14 35 0.90
CLA4_CANAL (O14427) Serine/threonine-protein kinase CLA4 (EC 2.7... 34 1.2
DEI_DROME (P41894) Helix-loop-helix protein delilah 33 2.0
CASR_BOVIN (P35384) Extracellular calcium-sensing receptor precu... 33 2.0
UTR2_YEAST (P32623) UTR2 protein (Unknown transcript 2 protein) 33 2.6
CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14) 33 2.6
SGS3_DROSI (P13729) Salivary glue protein Sgs-3 precursor 33 3.4
DF24_ARATH (O80928) Dof zinc finger protein DOF2.4 (AtDOF2.4) 33 3.4
CYAA_DICDI (Q03100) Adenylate cyclase, aggregation specific (EC ... 33 3.4
BOB1_YEAST (P38041) BOB1 protein (BEM1-binding protein) 33 3.4
>YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I
Length = 1261
Score = 55.1 bits (131), Expect = 6e-07
Identities = 66/275 (24%), Positives = 110/275 (40%), Gaps = 17/275 (6%)
Query: 228 YSTIISCAKKCNLFDKAVYWFE------RMYKTGLMPDEVTFSA-ILDVY---ARLGKVE 277
Y T+IS K FD FE R T + F A ILD + +
Sbjct: 810 YPTLISVLSKNKRFDAVQRVFEHSKHLYRKISTKSLEKANWFMALILDAMILSSSFARQF 869
Query: 278 EVVNLF-ERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVL---QEMKSLGVQPNLVVY 333
+ NLF + + G+ P TF+ L GD D L +E K V+P++ +Y
Sbjct: 870 KSSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSVFLY 929
Query: 334 NTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMK- 392
N +L +G+A + LF+EM +SG+ P T VI + A +L+ M+
Sbjct: 930 NAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAACRIGDESLAEKLFAEMEN 989
Query: 393 ENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAV 452
+ + YNT++ E + + + +P S +Y +++ YG+ V
Sbjct: 990 QPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCATDIEPSSHTYKLLMDAYGTLKPV 1049
Query: 453 D--KAMKLFEEMSKFGIELNVMGCTCLIQCLGKAM 485
+ + E M + + + M I LG +
Sbjct: 1050 NVGSVKAVLELMERTDVPILSMHYAAYIHILGNVV 1084
Score = 43.5 bits (101), Expect = 0.002
Identities = 60/297 (20%), Positives = 116/297 (38%), Gaps = 50/297 (16%)
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEE 211
PS L L R + F +K LLP + V + R G + + E+
Sbjct: 924 PSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAACRIGDE-SLAEK 982
Query: 212 LAHQMIDGGVELDNIT-YSTIISCAKKCNLF--DKAVYWFERMYKTGLMPDEVTFSAILD 268
L +M + + Y+T+I + +F +KA++++ R+ T + P T+ ++D
Sbjct: 983 LFAEMENQPNYQPRVAPYNTMIQFEVQ-TMFNREKALFYYNRLCATDIEPSSHTYKLLMD 1041
Query: 269 VYARL-----GKVEEVVNLFERG------------------------RATGWKPDPITFS 299
Y L G V+ V+ L ER AT + +
Sbjct: 1042 AYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHILGNVVSDVQAATSCYMNALAKH 1101
Query: 300 VLGKMFGEAGDY--------------DGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGK 345
G++ +A + +GI+ ++ +MK V N + N L++ K G
Sbjct: 1102 DAGEIQLDANLFQSQIESLIANDRIVEGIQ-IVSDMKRYNVSLNAYIVNALIKGFTKVGM 1160
Query: 346 PGFARSLFEEMIDSGIAPNE-KTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFI 401
AR F+ + G++ E T +++ Y + A+E+ +++K +P+ +
Sbjct: 1161 ISKARYYFDLLECEGMSGKEPSTYENMVRAYLSVNDGRKAMEIVEQLKRKRYPLPVV 1217
Score = 42.7 bits (99), Expect = 0.003
Identities = 69/330 (20%), Positives = 124/330 (36%), Gaps = 76/330 (23%)
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAK 236
F K N+ P YN + L R+ +L +M + G+ ++TY T+I+ A
Sbjct: 914 FEETKRHNVKP-SVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAAC 972
Query: 237 KC-------NLF------------------------------DKAVYWFERMYKTGLMPD 259
+ LF +KA++++ R+ T + P
Sbjct: 973 RIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCATDIEPS 1032
Query: 260 EVTFSAILDVYARL-----GKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGI 314
T+ ++D Y L G V+ V+ L ER PI + Y
Sbjct: 1033 SHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDV------PIL----------SMHYAAY 1076
Query: 315 RYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKI 374
++L + S VQ Y L A AG+ +LF+ I+S IA N++ +
Sbjct: 1077 IHILGNVVS-DVQAATSCYMNAL-AKHDAGEIQLDANLFQSQIESLIA-NDRIV------ 1127
Query: 375 YGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKP 434
+ +++ MK ++ + N L+ VG+I +A F ++
Sbjct: 1128 --------EGIQIVSDMKRYNVSLNAYIVNALIKGFTKVGMISKARYYFDLLECEGMSGK 1179
Query: 435 DSWSYTAMLNIYGSEGAVDKAMKLFEEMSK 464
+ +Y M+ Y S KAM++ E++ +
Sbjct: 1180 EPSTYENMVRAYLSVNDGRKAMEIVEQLKR 1209
Score = 32.3 bits (72), Expect = 4.5
Identities = 37/179 (20%), Positives = 74/179 (40%), Gaps = 9/179 (5%)
Query: 389 KRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGS 448
K +++ W M IL +L+ + ++ LF D + P + ++ ++N
Sbjct: 843 KSLEKANWFMALILDAMILS--SSFARQFKSSNLFCDNMKMLGYIPRASTFAHLINNSTR 900
Query: 449 EGAVDKA---MKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
G D A + +FEE + ++ +V ++ LG+A + K+F E G+ P
Sbjct: 901 RGDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPT 960
Query: 506 DRLCGCLLSVVSLSQGSKDQEKVLACLQRA---NPKLVAFIQLIVDE-ETSFETVKEEF 560
G +++ EK+ A ++ P++ + +I E +T F K F
Sbjct: 961 SVTYGTVINAACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALF 1019
>PT09_YEAST (P32522) PET309 protein, mitochondrial precursor
Length = 965
Score = 53.1 bits (126), Expect = 2e-06
Identities = 50/209 (23%), Positives = 86/209 (40%), Gaps = 5/209 (2%)
Query: 259 DEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVL 318
D+ F +L ++RL + + F G P + +L G+ D + +
Sbjct: 310 DQNQFDYLLVAHSRLHNWDALQQQFNALFGIGKLPSIQHYGILMYTMARIGELDSVNKLY 369
Query: 319 QEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKA 378
++ G+ P V +LL A K G S FE I P+ T T ++K+Y
Sbjct: 370 TQLLRRGMIPTYAVLQSLLYAHYKVGDFAACFSHFELFKKYDITPSTATHTIMLKVYRGL 429
Query: 379 RWSKDALELWKRMKENGWPMDFIL---YNTLLNMCADVGLIEEAETLFRDMKQSEHCKPD 435
A + KR+ E+ P I + L+ MC A+ LF M + + +
Sbjct: 430 NDLDGAFRILKRLSED--PSVEITEGHFALLIQMCCKTTNHLIAQELFNLMTEHYNIQHT 487
Query: 436 SWSYTAMLNIYGSEGAVDKAMKLFEEMSK 464
S +A++++Y +A+ LFE+ SK
Sbjct: 488 GKSISALMDVYIESNRPTEAIALFEKHSK 516
>LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP130)
(Leucine-rich PPR-motif containing protein)
Length = 1273
Score = 48.9 bits (115), Expect = 5e-05
Identities = 36/184 (19%), Positives = 72/184 (38%), Gaps = 4/184 (2%)
Query: 211 ELAHQMIDG----GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAI 266
E AH++ D G D Y+ ++ + + +M + + P+ VT+ +
Sbjct: 23 EFAHRIWDTLQKLGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRL 82
Query: 267 LDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGV 326
+ Y +G +E + + FS L AGD + +L M+ G+
Sbjct: 83 IASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGI 142
Query: 327 QPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALE 386
+P Y LL A + G + E++ + ++ L +I + KA + + +
Sbjct: 143 EPGPDTYLALLNAYAEKGDIDHVKQTLEKVEKFELHLMDRDLLQIIFSFSKAGYLSMSQK 202
Query: 387 LWKR 390
WK+
Sbjct: 203 FWKK 206
Score = 48.5 bits (114), Expect = 6e-05
Identities = 36/149 (24%), Positives = 67/149 (44%), Gaps = 1/149 (0%)
Query: 321 MKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARW 380
++ LG ++ YN LL+ + +M ++ I PN T +I Y
Sbjct: 32 LQKLGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGD 91
Query: 381 SKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYT 440
+ A ++ MK P+ +++ L+ A G +E AE + M+ + +P +Y
Sbjct: 92 IEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAG-IEPGPDTYL 150
Query: 441 AMLNIYGSEGAVDKAMKLFEEMSKFGIEL 469
A+LN Y +G +D + E++ KF + L
Sbjct: 151 ALLNAYAEKGDIDHVKQTLEKVEKFELHL 179
Score = 47.4 bits (111), Expect = 1e-04
Identities = 42/159 (26%), Positives = 76/159 (47%), Gaps = 9/159 (5%)
Query: 193 YNVTMKS-LRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFD-KAVYWFER 250
YN +K L+ +F + LA +M + ++ + +TY +I A CN+ D +
Sbjct: 44 YNALLKVYLQNEYKFSPTDFLA-KMEEANIQPNRVTYQRLI--ASYCNVGDIEGASKILG 100
Query: 251 MYKTGLMP-DEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
KT +P E FSA++ +AR G +E N+ R G +P P T+ L + E G
Sbjct: 101 FMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNAYAEKG 160
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGF 348
D D ++ L++++ + ++ LL+ + K G+
Sbjct: 161 DIDHVKQTLEKVEKFELH---LMDRDLLQIIFSFSKAGY 196
>YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR1
intergenic region
Length = 864
Score = 47.0 bits (110), Expect = 2e-04
Identities = 30/97 (30%), Positives = 48/97 (48%), Gaps = 5/97 (5%)
Query: 358 DSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMK---ENGWPMDFILYNTLLNMCADVG 414
D GI PN++ LT VI+ Y + +K A + MK +P D YNT+L +C
Sbjct: 313 DYGITPNKQNLTTVIQFYSRKEMTKQAWNTFDTMKFLSTKHFP-DICTYNTMLRICEKER 371
Query: 415 LIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
+A LF+++ Q + KP + +Y M + S +
Sbjct: 372 NFPKALDLFQEI-QDHNIKPTTNTYIMMARVLASSSS 407
>DAN4_YEAST (P47179) Cell wall protein DAN4 precursor
Length = 1161
Score = 41.6 bits (96), Expect = 0.007
Identities = 45/174 (25%), Positives = 74/174 (41%), Gaps = 11/174 (6%)
Query: 2 ATSLSSSIHISFLDTKTTRTRFKFPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKK 61
+++ +SS+ + T TT P T+T H +S++ + S TT ++
Sbjct: 303 SSASASSVISTTATTSTTFASLTTPATSTASTDHTTSSVSTTNAFTTSATTTTTSDTYIS 362
Query: 62 NSSLSDQLASLANTTLSTV-----PENQPKVLSKPKPTWVNP-TKTKRPVLSHQRHKRSS 115
+SS S +S TT+S V P +V S +PT V+ T + P S Q +S
Sbjct: 363 SSSPSQVTSSAEPTTVSEVTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPTRSSQ---VTS 419
Query: 116 VSYNPQLREFQRFAQRLNNCDVSSSDEEFMVCLEEIPSSLTRGNALLVLNSLRP 169
+ + EF + + V+SS E V E SS+ + V +S P
Sbjct: 420 SAEPTTVSEFTSSVEPTRSSQVTSSAEPTTV--SEFTSSVEPTRSSQVTSSAEP 471
>NPR1_YEAST (P22211) Nitrogen permease reactivator protein (EC
2.7.1.-)
Length = 790
Score = 37.0 bits (84), Expect = 0.18
Identities = 39/130 (30%), Positives = 60/130 (46%), Gaps = 24/130 (18%)
Query: 29 TTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLSTVPENQP--- 85
+TT + R + S S SIP S N+NK N+S + +SL+++ L T P
Sbjct: 153 STTSHTSGRAIPSLSSSIPYSVP-----NSNKDNNSSNSNSSSLSSSWLETYAGGMPNNI 207
Query: 86 -----KVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYN----PQLREFQR---FAQRLN 133
V+S PK V P R V+S Q+ +++S+ N Q R R F+ +L
Sbjct: 208 SAIDSNVISSPKVDSVEP----RFVISKQKLQKASMDSNNANATQSRSISRSGSFSSQLG 263
Query: 134 NCDVSSSDEE 143
N S + +E
Sbjct: 264 NFFFSKNSKE 273
>PTPX_CANAL (P43078) Probable protein-tyrosine phosphatase (EC
3.1.3.48)
Length = 597
Score = 36.6 bits (83), Expect = 0.24
Identities = 20/44 (45%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Query: 40 ISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLSTVPEN 83
++SS SI + E T NN NNK N++ S ++ NT LST P+N
Sbjct: 360 LNSSFSINNDEATNFNNKNNKNNNNNSTATTTITNTILST-PQN 402
>A3B1_MOUSE (Q9Z1T1) Adapter-related protein complex 3 beta 1
subunit (Beta3A-adaptin) (Adaptor protein complex AP-3
beta-1 subunit) (AP-3 complex beta-1 subunit) (Clathrin
assembly protein complex 3 beta-1 large chain)
Length = 1105
Score = 35.8 bits (81), Expect = 0.40
Identities = 25/85 (29%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query: 41 SSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTWVNPTK 100
S +K DSE N N+K + S S++ +S+ ++ S+ E++ S+P P V P K
Sbjct: 755 SKTKRKSDSENREKKNENSKASESSSEESSSMEDS--SSESESESGSDSEPAPRNVAPAK 812
Query: 101 TKRPVLSHQRHKRSSVSYNPQLREF 125
++P +RH S + L +F
Sbjct: 813 ERKP--QQERHPPSKDVFLLDLDDF 835
>SSD1_YEAST (P24276) SSD1 protein (SRK1 protein)
Length = 1250
Score = 35.4 bits (80), Expect = 0.53
Identities = 29/106 (27%), Positives = 49/106 (45%), Gaps = 10/106 (9%)
Query: 15 DTKTTRTRFKFPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLAN 74
++ T ++ FKFPP + HRR ++S P S PPN++ + + ++ +
Sbjct: 203 NSTTEQSDFKFPPPPNAHQGHRR---ATSNLSPPSFKFPPNSHGDNDDEFIATSSTHRRS 259
Query: 75 TTLSTVPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRS-SVSYN 119
T + E P + S W N ++ + LS RH+ S S YN
Sbjct: 260 KTRNN--EYSPGINS----NWRNQSQQPQQQLSPFRHRGSNSRDYN 299
>TKT_BUCAP (Q8KA26) Transketolase (EC 2.2.1.1) (TK)
Length = 665
Score = 34.7 bits (78), Expect = 0.90
Identities = 35/134 (26%), Positives = 56/134 (41%), Gaps = 22/134 (16%)
Query: 286 GRATGWKPDPIT-------FSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLE 338
G+ + W D + V+ K+ G D + I+ ++E KS+ QP++++ NT++
Sbjct: 191 GKISNWFTDDTVMRFKSYNWHVVDKVDGH--DANSIKNSIEEAKSVKDQPSIIICNTII- 247
Query: 339 AMGKAGKPGFARSLFEEMIDSGIAPNEKTLT-------AVIKIYGKARWSKDALELWKRM 391
G K G A S + + I K L KIY K + K L+L
Sbjct: 248 GFGSPNKSGTADSHGAPLGEEEIFLTRKNLNWKYSPFEIPNKIYDKWNFVKQGLKL---- 303
Query: 392 KENGWPMDFILYNT 405
E W F LY +
Sbjct: 304 -EENWNKQFHLYKS 316
>SX14_DROME (P40656) Putative transcription factor SOX-14
Length = 472
Score = 34.7 bits (78), Expect = 0.90
Identities = 39/151 (25%), Positives = 57/151 (36%), Gaps = 12/151 (7%)
Query: 26 PPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQLASL--ANTTLSTVPEN 83
P T+ RR + S P T PP + + SS S A A T S
Sbjct: 29 PARPATITIQRRHPAPKADSTP--HTLPPFSPSPSPASSPSPAPAQTPGAQKTQSQAAIT 86
Query: 84 QPKVLSKPKPTWV----NPTKTKRPVLSHQRHKRSSVSYNPQLREFQRFAQRLNNCDVSS 139
P ++ P KT P +H S ++P RE + +R S
Sbjct: 87 HPAAVASPSAPVAAAAPKTPKTPEPRSTHTHTHTHSQHFSPPPRESEMDGER----SPSH 142
Query: 140 SDEEFMVCLEEIPSSLTRGNALLVLNSLRPW 170
S E + ++ I SSL G+A + +NS P+
Sbjct: 143 SGHEMTLSMDGIDSSLVFGSARVPVNSSTPY 173
>CLA4_CANAL (O14427) Serine/threonine-protein kinase CLA4 (EC
2.7.1.37)
Length = 971
Score = 34.3 bits (77), Expect = 1.2
Identities = 33/127 (25%), Positives = 51/127 (39%), Gaps = 13/127 (10%)
Query: 17 KTTRTRFK------FPPTT---TTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSD 67
K+T ++FK PPT T L SSS S+P S NN+ N N+
Sbjct: 341 KSTVSQFKPSRAAPKPPTPYHLTQLNGSSHQHTSSSGSLPSSGNNNNNNSTNNNNTKNVS 400
Query: 68 QLASLANTTLSTVPENQ---PKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYNPQLRE 124
L +L N + +P + P T+ N R QR +R+ S Q ++
Sbjct: 401 PLNNLMNKS-ELIPARRAPPPPTSGTSSDTYSNKNHQDRSGYEQQRQQRTDSSQQQQQQK 459
Query: 125 FQRFAQR 131
++ Q+
Sbjct: 460 QHQYQQK 466
>DEI_DROME (P41894) Helix-loop-helix protein delilah
Length = 360
Score = 33.5 bits (75), Expect = 2.0
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 2/76 (2%)
Query: 60 KKNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYN 119
K+ S+ + A+LA+ L P +P SK + +KTK P LS R K ++
Sbjct: 47 KRGSNEDGESANLADFQLELDPIAEPA--SKSRKNAPTKSKTKAPPLSKYRRKTANARER 104
Query: 120 PQLREFQRFAQRLNNC 135
++RE + L +C
Sbjct: 105 TRMREINTAFETLRHC 120
>CASR_BOVIN (P35384) Extracellular calcium-sensing receptor
precursor (CaSR) (Parathyroid Cell calcium-sensing
receptor)
Length = 1085
Score = 33.5 bits (75), Expect = 2.0
Identities = 25/115 (21%), Positives = 53/115 (45%), Gaps = 11/115 (9%)
Query: 25 FPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLANTTLST----- 79
F P+ T++ R + + + T +N + +++SSL S ++++S+
Sbjct: 863 FKPSRNTIEEVRCSTAAHAFKVAARATLRRSNVSRQRSSSLGGSTGSTPSSSISSKSNSE 922
Query: 80 --VPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYNPQLREFQRFAQRL 132
P+ QPK +P+P ++P ++P Q S+ PQ ++ R Q++
Sbjct: 923 DPFPQQQPKRQKQPQPLALSPHNAQQP----QPRPPSTPQPQPQSQQPPRCKQKV 973
>UTR2_YEAST (P32623) UTR2 protein (Unknown transcript 2 protein)
Length = 347
Score = 33.1 bits (74), Expect = 2.6
Identities = 28/98 (28%), Positives = 49/98 (49%), Gaps = 12/98 (12%)
Query: 28 TTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNS------SLSDQLASLANTTLSTVP 81
T++T KS SSS + S+T+ ++++ KK+S S S +S ++++ ST
Sbjct: 233 TSSTQKS------SSSTATSSSKTSSDHSSSTKKSSKTSSTASSSSSSSSSSSSSSSTAT 286
Query: 82 ENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYN 119
+N KV+S + + T+T V SS+S N
Sbjct: 287 KNGDKVVSSVSSSVTSQTQTTSSVSGSASSSTSSMSGN 324
>CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14)
Length = 567
Score = 33.1 bits (74), Expect = 2.6
Identities = 29/106 (27%), Positives = 55/106 (51%), Gaps = 9/106 (8%)
Query: 2 ATSLSSSIHISFLDTKTTRTRFKFPPTTTTLKSHRRFLISSSKSIP-DSETTPPNNNNNK 60
+TSLSSS + T T + T S +SS+ +IP DS TT + +++
Sbjct: 400 STSLSSSTISTSASTSDTTSVTSSETTPVVTPSS----LSSAITIPGDSTTTGISKSSST 455
Query: 61 KNSSLSDQLASLANTTLSTVPENQPKVLSKPKPTWVNPTKTKRPVL 106
K ++ + S + TT++T+P+++ ++++ P T T +K P +
Sbjct: 456 KPATSTTSALSSSTTTVATIPDDK-EIINTPTDT---ETTSKPPAI 497
>SGS3_DROSI (P13729) Salivary glue protein Sgs-3 precursor
Length = 217
Score = 32.7 bits (73), Expect = 3.4
Identities = 21/73 (28%), Positives = 29/73 (38%), Gaps = 6/73 (8%)
Query: 16 TKTTRTRFKFPPTTT------TLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQL 69
T TT T + PPTTT T +H+ S+ K+ P TTP
Sbjct: 95 TSTTTTTTRAPPTTTCKTSTTTTTTHKPTTHSTPKTKPTKHTTPKTKPTKHTTPKTKPTK 154
Query: 70 ASLANTTLSTVPE 82
+ TT +T P+
Sbjct: 155 HTTPTTTTTTTPK 167
>DF24_ARATH (O80928) Dof zinc finger protein DOF2.4 (AtDOF2.4)
Length = 330
Score = 32.7 bits (73), Expect = 3.4
Identities = 28/103 (27%), Positives = 45/103 (43%), Gaps = 11/103 (10%)
Query: 55 NNNNNK----KNSSLSDQLASLANTTLST-VPENQPKVLSKPKPTWVNPTKTKRPVLSH- 108
NNNNN N+S S AS +T LS+ NQ +LS+ ++P + P +H
Sbjct: 149 NNNNNSTATSNNTSFSSGNASTISTILSSHYGGNQESILSQ----ILSPARLMNPTYNHL 204
Query: 109 -QRHKRSSVSYNPQLREFQRFAQRLNNCDVSSSDEEFMVCLEE 150
+ N L + +Q L + + +S M C++E
Sbjct: 205 GDLTSNTKTDNNMSLLNYGGLSQDLRSIHMGASGGSLMSCVDE 247
>CYAA_DICDI (Q03100) Adenylate cyclase, aggregation specific (EC
4.6.1.1) (ATP pyrophosphate-lyase) (Adenylyl cyclase)
Length = 1407
Score = 32.7 bits (73), Expect = 3.4
Identities = 29/118 (24%), Positives = 44/118 (36%), Gaps = 39/118 (33%)
Query: 47 PDSETTPPNNNNNKKNSSLSDQ----------------------LASLANTTLSTVPENQ 84
P++ +TP NNN+N N S S LA LAN ++
Sbjct: 98 PNTLSTPHNNNHNNNNHSTSHHPHSNSVANGGHLSQSITQQRGGLADLANAVINR-KNRS 156
Query: 85 PKVLSKPKPT--------WVNPTKTKRPVLSHQRHKRSSVSYNPQLREFQRFAQRLNN 134
V +K KPT W K + ++ K+S++ FQ++ RL N
Sbjct: 157 DSVQTKMKPTDSASNIESWAKVEKFSSSIFDSEKSKKSNI--------FQKYTLRLKN 206
>BOB1_YEAST (P38041) BOB1 protein (BEM1-binding protein)
Length = 980
Score = 32.7 bits (73), Expect = 3.4
Identities = 28/111 (25%), Positives = 49/111 (43%), Gaps = 6/111 (5%)
Query: 15 DTKTTRTRFKFPPTTTTLKSHRRFLISSSKSIPDSETTPPNNNNNKKNSSLSDQLASLAN 74
+ + T +FPP TT ++ + S+ IP+S T+ N+ K +++ ++
Sbjct: 416 NNRYTNNNARFPPQTTYPPKNKNPTVYSNGLIPNSSTSSDNSTGKFKFPAMNGHDSNSRK 475
Query: 75 TTL--STVPENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYNPQLR 123
TTL +T+P + P N + SH + R+SV YN R
Sbjct: 476 TTLTSATIPSINTVNTDESLPAISNISSN---ATSHHPN-RNSVVYNNHKR 522
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 82,310,408
Number of Sequences: 164201
Number of extensions: 3489595
Number of successful extensions: 10667
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 10533
Number of HSP's gapped (non-prelim): 118
length of query: 715
length of database: 59,974,054
effective HSP length: 117
effective length of query: 598
effective length of database: 40,762,537
effective search space: 24375997126
effective search space used: 24375997126
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146791.8


