

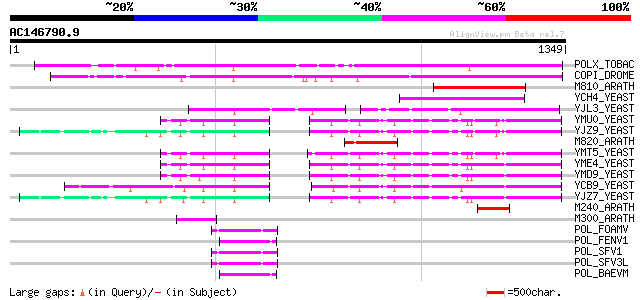
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.9 + phase: 0
(1349 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 564 e-160
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 540 e-152
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 213 2e-54
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 161 1e-38
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 127 2e-28
YMU0_YEAST (Q04670) Transposon Ty1 protein B 124 2e-27
YJZ9_YEAST (P47100) Transposon Ty1 protein B 122 5e-27
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 122 7e-27
YMT5_YEAST (Q04214) Transposon Ty1 protein B 120 2e-26
YME4_YEAST (Q04711) Transposon Ty1 protein B 120 2e-26
YMD9_YEAST (Q03434) Transposon Ty1 protein B 120 3e-26
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 118 1e-25
YJZ7_YEAST (P47098) Transposon Ty1 protein B 118 1e-25
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 76 5e-13
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 58 2e-07
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 52 8e-06
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 52 1e-05
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 50 3e-05
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 50 4e-05
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 50 4e-05
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 564 bits (1453), Expect = e-160
Identities = 416/1340 (31%), Positives = 648/1340 (48%), Gaps = 110/1340 (8%)
Query: 60 DSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQ 119
DS K + E+W D+R + ++ ++ ++ +T++ +W +SL + T +
Sbjct: 40 DSKKPDTMKAEDWADLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTN 99
Query: 120 IIYLKFQFHSIRKGE-MKMEDYLIKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPI 178
+YLK Q +++ E +L L +L G I + I LN L S Y+ +
Sbjct: 100 KLYLKKQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNL 159
Query: 179 VVK-LSDHTTLSWVDLQAQLLTFESRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNN 237
L TT+ D+ + LL LN N+ +
Sbjct: 160 ATTILHGKTTIELKDVTSALL--------------LNEKMRKKPENQGQALITEGRGRSY 205
Query: 238 WRGSNFRGWRGGRGRGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHN 297
R SN G G RG+ ++ ++ +V N F R N + K +
Sbjct: 206 QRSSNNYGRSGARGKSKN-RSKSRVRNCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNT 264
Query: 298 AFIASQN----------------SVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQV 341
A + N S + +W D+ AS+H T D F G V
Sbjct: 265 AAMVQNNDNVVLFINEEEECMHLSGPESEWVVDTAASHHATPVRDLFCRYVA--GDFGTV 322
Query: 342 -VGNGDKLEIVATGSSKLKS-----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
+GN +I G +K+ L L DV +VP++ NL +S +A D + + + N
Sbjct: 323 KMGNTSYSKIAGIGDICIKTNVGCTLVLKDVRHVPDLRMNL--ISGIALDRDGYESYFAN 380
Query: 396 CCFVKEKLT--GKVILKGLLKNGLYQLSDT--KGNPYAF---VSVKESWHRRLGHPNNKV 448
K +LT VI KG+ + LY+ + +G A +SV + WH+R+GH + K
Sbjct: 381 ---QKWRLTKGSLVIAKGVARGTLYRTNAEICQGELNAAQDEISV-DLWHKRMGHMSEKG 436
Query: 449 LDKVLKSCNVKVPPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWGPAPIMS 508
L + K + C+ C +GK H + F++SS L+LV++DV GP I S
Sbjct: 437 LQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIES 496
Query: 509 SSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFT-----TENQFNKRIKVIQCDGGGEY- 562
G KY+V FIDD SR W+Y LK K + Q F E + +++K ++ D GGEY
Sbjct: 497 MGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYT 556
Query: 563 -KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFST 621
+ ++ GI+ + P T Q NG AER +R I E ++L A++P +W EA T
Sbjct: 557 SREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQT 616
Query: 622 AVYLINRLPSQVTQNESPYSLIFHKEPNYKLLKPFGCACYPCLKPYNQHKLQFHTTRCVF 681
A YLINR PS E P + +KE +Y LK FGC + + + KL + C+F
Sbjct: 617 ACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIF 676
Query: 682 LGYSNSHKGYKCLNSHGRTFISRHVIFNEDLFPFHEGFLNTRSPLKTTINNPSTSFPLCS 741
+GY + GY+ + + + VI + D+ F E + T + + + N +
Sbjct: 677 IGYGDEEFGYRLWDP-----VKKKVIRSRDVV-FRESEVRTAADMSEKVKNGIIPNFVTI 730
Query: 742 AGNSINDASIPIIEEENQDETNEEDSQG--VTSDTEQTDNGSSEGD--TTHEETLDIVQQ 797
S N P E DE +E+ Q V EQ D G E + T EE Q
Sbjct: 731 PSTSNN----PTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEE-----QH 781
Query: 798 QNVGESSLDTNTSNAIHTRSKSGIHKPKLPYIGITETYKDTVEPANVKEALT---RTLWK 854
Q + S S P Y+ I+ D EP ++KE L+ +
Sbjct: 782 QPLRRSERPRVESRRY----------PSTEYVLIS----DDREPESLKEVLSHPEKNQLM 827
Query: 855 EAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTA 914
+AMQ+E ++L N T+ LV + + KWVFK K D + R KARLV KGF+Q
Sbjct: 828 KAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKK 887
Query: 915 GIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDP 974
GID++E FSPVVK++++R ILS+A L+ EV QLD+ AFL+G L+E ++M QPEGF
Sbjct: 888 GIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVA 947
Query: 975 TKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLK-GKDHITFLLI 1033
K + +CKL+K++YGLKQAPR W+ + + + + T SDP ++ + +++ LL+
Sbjct: 948 GKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLL 1007
Query: 1034 YVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASG--MYLKQSKYIG 1091
YVDD+++ G + L+ F +KDLG LG+++ R+ + ++L Q KYI
Sbjct: 1008 YVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIE 1067
Query: 1092 DLLKKFKMENASPCPTPMITGRHF-------TVEGEKLKDPTVFRQAIGGLQY-LTHTRP 1143
+L++F M+NA P TP+ TVE + + A+G L Y + TRP
Sbjct: 1068 RVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRP 1127
Query: 1144 DIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATS 1203
DIA +V +S+++ +P +HW+ +K ILRYL+GT CL S D + G++DAD A
Sbjct: 1128 DIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGS-DPILKGYTDADMAGD 1186
Query: 1204 IDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKL 1263
ID+RKS G ISW S+ QK V+ S+TE+EY A + E+ W+ L EL L
Sbjct: 1187 IDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGL 1246
Query: 1264 PLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIAD 1323
++ +++CD+ SA L+ N + HAR+KHI++ H+IR+ V + V + T + AD
Sbjct: 1247 H-QKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPAD 1305
Query: 1324 CLTKPLSHTRFSQLRDKLGV 1343
LTK + +F ++ +G+
Sbjct: 1306 MLTKVVPRNKFELCKELVGM 1325
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 540 bits (1391), Expect = e-152
Identities = 403/1352 (29%), Positives = 666/1352 (48%), Gaps = 133/1352 (9%)
Query: 100 TSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIR-KGEMKMEDYLIKMKNLADKLKLAGNPI 158
T++Q+ + ++ + + + L+ + S++ EM + + L +L AG I
Sbjct: 76 TARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELISELLAAGAKI 135
Query: 159 SNSNLIIQTLNGLDSEYNPIVVKLS----DHTTLSWVDLQAQLLTFESRIEQLNTLTNLN 214
+ I L L S Y+ I+ + ++ TL++V + +LL E +I+ N +
Sbjct: 136 EEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFV--KNRLLDQEIKIK------NDH 187
Query: 215 LNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQVYGKTNHTAINCF 274
+ + V N H +N NN ++ + + +G + K C G+ H +CF
Sbjct: 188 NDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKY-KVKCHHCGREGHIKKDCF 246
Query: 275 HRFDKNYSR--SNYSADSDKQ----GSHN-AFIASQ----NSVEDYDWYFDSGASNHVTH 323
H Y R +N + +++KQ SH AF+ + + +++ + DSGAS+H+ +
Sbjct: 247 H-----YKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGASDHLIN 301
Query: 324 QTDKFQDLTEHHGK-NSQVVGNGDKLEIVATGSSKLKS---LNLDDVLYVPNITKNLLSV 379
+ D E V G+ + G +L++ + L+DVL+ NL+SV
Sbjct: 302 DESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEITLEDVLFCKEAAGNLMSV 361
Query: 380 SKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKN------GLYQLSDTKGNPYAFVSV 433
+L + + +EFDK+ + + V G+L N Y ++ N +
Sbjct: 362 KRLQ-EAGMSIEFDKSGVTISKNGLMVVKNSGMLNNVPVINFQAYSINAKHKNNFRL--- 417
Query: 434 KESWHRRLGH-PNNKVLDKVLKSCNVKVPPSDNFSF----CEACQYGKMHLLPFKS--SS 486
WH R GH + K+L+ K+ +N CE C GK LPFK
Sbjct: 418 ---WHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQARLPFKQLKDK 474
Query: 487 SHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFT---- 542
+H + PL +VH+DV GP ++ Y+V F+D F+ + Y +K KS+ F
Sbjct: 475 THIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVA 534
Query: 543 -TENQFNKRIKVIQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAE 599
+E FN ++ + D G EY +++ + GI + ++ P+T Q NG +ER R I E
Sbjct: 535 KSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITE 594
Query: 600 FGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQ--VTQNESPYSLIFHKEPNYKLLKPFG 657
T+++ A++ +W EA TA YLINR+PS+ V +++PY + +K+P K L+ FG
Sbjct: 595 KARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFG 654
Query: 658 CACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLNSHGRTFI-SRHVIFNED----- 711
Y +K Q K + + +F+GY + G+K ++ FI +R V+ +E
Sbjct: 655 ATVYVHIKN-KQGKFDDKSFKSIFVGYEPN--GFKLWDAVNEKFIVARDVVVDETNMVNS 711
Query: 712 -LFPFHEGFL-------------NTRSPLKTTINNPSTSFPLC--------SAGNSINDA 749
F FL ++R ++T N S S + +
Sbjct: 712 RAVKFETVFLKDSKESENKNFPNDSRKIIQTEFPNESKECDNIQFLKDSKESENKNFPND 771
Query: 750 SIPIIEEENQDETNEEDSQGVTSDTEQTDN---------------GSSEGDTTHEETLDI 794
S II+ E +E+ E D+ D+++++ S+G E+ +
Sbjct: 772 SRKIIQTEFPNESKECDNIQFLKDSKESNKYFLNESKKRKRDDHLNESKGSGNPNESRES 831
Query: 795 VQQQNVGESSLDTNTSN----AIHTRSKSGIHKPKLPYIGITETYKDTVEPAN------- 843
+++ E +D T N I+ RS+ KP++ Y + V A+
Sbjct: 832 ETAEHLKEIGIDNPTKNDGIEIINRRSERLKTKPQISYNEEDNSLNKVVLNAHTIFNDVP 891
Query: 844 -----VKEALTRTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKYKSDGSI 898
++ ++ W+EA+ E A N TW + + +NIVDS+WVF KY G+
Sbjct: 892 NSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNP 951
Query: 899 ERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRVILSIAVHLNWEVRQLDINNAFLNGY 958
R KARLVA+GF Q IDYEETF+PV ++S+ R ILS+ + N +V Q+D+ AFLNG
Sbjct: 952 IRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGT 1011
Query: 959 LKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPS 1018
LKE ++M P+G +++CKL+KAIYGLKQA R WF+ + AL F N+ D
Sbjct: 1012 LKEEIYMRLPQGI--SCNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRC 1069
Query: 1019 LFLL-KGK-DHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQ 1076
+++L KG + ++L+YVDD+++ + F + L + F + DL + +F+GI ++
Sbjct: 1070 IYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIE 1129
Query: 1077 RDASGMYLKQSKYIGDLLKKFKMENASPCPTPMITGRHFTVEGEKLKDPTVFRQAIGGLQ 1136
+YL QS Y+ +L KF MEN + TP+ + ++ + T R IG L
Sbjct: 1130 MQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYELLNSDEDCNTPCRSLIGCLM 1189
Query: 1137 Y-LTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLD--IT 1193
Y + TRPD+ +VN LS+Y S ++ WQ +KR+LRYL+GTI+ L K + + I
Sbjct: 1190 YIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKII 1249
Query: 1194 GFSDADWATSIDDRKSMAGQCVFLGE-TLISWSSRKQKVVSRSSTESEYRALVDLAAEIA 1252
G+ D+DWA S DRKS G + + LI W++++Q V+ SSTE+EY AL + E
Sbjct: 1250 GYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREAL 1309
Query: 1253 WIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVV 1312
W+ LL + + L ++ DN ++A+NP H R+KHI+I H+ R+QV N + +
Sbjct: 1310 WLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICL 1369
Query: 1313 AYVPTTDQIADCLTKPLSHTRFSQLRDKLGVI 1344
Y+PT +Q+AD TKPL RF +LRDKLG++
Sbjct: 1370 EYIPTENQLADIFTKPLPAARFVELRDKLGLL 1401
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 213 bits (543), Expect = 2e-54
Identities = 106/224 (47%), Positives = 149/224 (66%)
Query: 1030 FLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMYLKQSKY 1089
+LL+YVDDI++TGSSN L I QL+ FS+KDLG +HYFLGI+++ SG++L Q+KY
Sbjct: 2 YLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKY 61
Query: 1090 IGDLLKKFKMENASPCPTPMITGRHFTVEGEKLKDPTVFRQAIGGLQYLTHTRPDIAFSV 1149
+L M + P TP+ + +V K DP+ FR +G LQYLT TRPDI+++V
Sbjct: 62 AEQILNNAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAV 121
Query: 1150 NKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFSDADWATSIDDRKS 1209
N + Q M PT + +KR+LRY++GTI + L+I ++ L++ F D+DWA R+S
Sbjct: 122 NIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRS 181
Query: 1210 MAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAW 1253
G C FLG +ISWS+++Q VSRSSTE+EYRAL AAE+ W
Sbjct: 182 TTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 161 bits (407), Expect = 1e-38
Identities = 92/308 (29%), Positives = 158/308 (50%), Gaps = 4/308 (1%)
Query: 948 LDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLN 1007
+D++ AFLN + E +++ QP GFV+ P+++ +L +YGLKQAP W + + L
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1008 WGFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRL 1067
GF + + L+ D ++ +YVDD++V S ++L ++S+KDLG++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1068 HYFLGIEVQRDASG-MYLKQSKYIGDLLKKFKMENASPCPTPMITGRH-FTVEGEKLKDP 1125
FLG+ + + +G + L YI + ++ TP+ + F LKD
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 1126 TVFRQAIGGLQYLTHT-RPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHI 1184
T ++ +G L + +T RPDI++ V+ LS+++ P H + +R+LRYL T + CL
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 1185 KPSTDLDITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQK-VVSRSSTESEYRA 1243
+ + + +T + DA D S G L ++WSS+K K V+ STE+EY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 1244 LVDLAAEI 1251
+ EI
Sbjct: 301 ASETVMEI 308
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 127 bits (320), Expect = 2e-28
Identities = 132/517 (25%), Positives = 227/517 (43%), Gaps = 55/517 (10%)
Query: 853 WKEAMQKEFQALMSNKTW-ILVPYQDQEN----IVDSKWVFKTKYKSDGSIERRKARLVA 907
+K+A KE Q L K + + V Y E IV + +F K KAR+V
Sbjct: 1289 YKQAYHKELQNLKDMKVFDVDVKYSRSEIPDNLIVPTNTIFTKKRNGI-----YKARIVC 1343
Query: 908 KGFQQTAGIDYEETFSPVVKVST----VRVILSIAVHLNWEVRQLDINNAFLNGYLKETV 963
+G Q+ +T+S + S +++ L IA + N ++ LDIN+AFL L+E +
Sbjct: 1344 RGDTQSP-----DTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEEEI 1398
Query: 964 FMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALLNWGFQNTKSDPSLFLLK 1023
++ P + KL+KA+YGLKQ+P+ W D L+ L G ++ P L+ +
Sbjct: 1399 YIPHPHD------RRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLYQTE 1452
Query: 1024 GKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLKDLGRLHYFLGIEVQRDASGMY 1083
K+ + +YVDD ++ S+ L FI +L F LK G L + + D GM
Sbjct: 1453 DKN--LMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTL---IDDVLDTDILGMD 1507
Query: 1084 LKQSKYIGDL------------------LKKFKMENASPCPTPMITGRHFTV---EGEKL 1122
L +K +G + LKK + + T I + + E E
Sbjct: 1508 LVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFR 1567
Query: 1123 KDPTVFRQAIGGLQYLTH-TRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYC 1181
+ +Q +G L Y+ H R DI F+V K+++ ++ P + I +I++YL +
Sbjct: 1568 QGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIG 1627
Query: 1182 LHIKPSTDLD--ITGFSDADWATSIDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTES 1239
+H + D + +DA + D +S G ++ G + + S K SSTE+
Sbjct: 1628 IHYDRDCNKDKKVIAITDASVGSEY-DAQSRIGVILWYGMNIFNVYSNKSTNRCVSSTEA 1686
Query: 1240 EYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVH 1299
E A+ + A+ + L EL ++ D+ A + + K I
Sbjct: 1687 ELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTE 1746
Query: 1300 YIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQ 1336
I++++ + + + + IAD LTKP+S + F +
Sbjct: 1747 IIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFKR 1783
Score = 66.6 bits (161), Expect = 4e-10
Identities = 92/414 (22%), Positives = 169/414 (40%), Gaps = 44/414 (10%)
Query: 435 ESWHRRLGHPNNKVLDKVLKSCNVK----VPPSDNFSFCEACQYGKM----HLLPFKSSS 486
E H+R+GH + ++ +K + + + N +C+ C+ K H ++
Sbjct: 559 EDAHKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNH 618
Query: 487 SHAQEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYP-LKQKSETVQAFTTEN 545
S EP D++GP ++ +Y + +D+ +R+ + +ET+ A +N
Sbjct: 619 STDHEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKN 678
Query: 546 ------QFNKRIKVIQCDGGGEYK--PVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHI 597
QF+++++ I D G E+ +++ I GI ++ NGRAER R I
Sbjct: 679 IQYVETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTI 738
Query: 598 AEFGLTLLAQAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPNYKLLK--P 655
TLL Q+ + + +W A ++A + N L + T + P I + +L+ P
Sbjct: 739 ITDATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKST-GKLPLKAISRQPVTVRLMSFLP 797
Query: 656 FGCACYPCLKPYNQHKLQFHTTRCVFLGYSNSHKGYKCLNSHGRTFISRHVIFNEDLFPF 715
FG + +N KL+ + L + GYK S++ I D +
Sbjct: 798 FGEK--GIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFF------IPSKNKIVTSDNYTI 849
Query: 716 HEGFLNTRSPLKTTINNP--------------STSFPLCSAGNSINDASIPIIEEENQDE 761
++ R IN T LC A + D + PI E+
Sbjct: 850 PNYTMDGRVRNTQNINKSHQFSSDNDDEEDQIETVTNLCEALENYEDDNKPITRLEDL-F 908
Query: 762 TNEEDSQGVTSDTEQTDNGSSEGDTTHEETLDIVQQQNVGESSLDTNTSNAIHT 815
T EE SQ ++ + + + EGD + + D+ + + S + T+N+I T
Sbjct: 909 TEEELSQIDSNAKYPSPSNNLEGDLDYVFS-DVEESGDYDVESELSTTNNSIST 961
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 124 bits (310), Expect = 2e-27
Identities = 164/672 (24%), Positives = 288/672 (42%), Gaps = 92/672 (13%)
Query: 730 INNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN-----GSSEG 784
I P+T + + I D +P + E+ E + + ++ QT++ G S
Sbjct: 685 IKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELPPINSRQTNSSLGGIGDSNA 744
Query: 785 DTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITETYKDTV 839
TT + + + + + S DT NT N RSK IH + + ++ K
Sbjct: 745 YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKSIKPIR 802
Query: 840 EPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY 892
EA+T + + +A KE L+ KTW Y D++ I D K V + +
Sbjct: 803 TTLRYDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEI-DPKRVINSMF 861
Query: 893 ----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNW 943
K DG+ KAR VA+G I + +T+ P ++ +TV LS+A+ N+
Sbjct: 862 IFNRKRDGT---HKARFVARG-----DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNY 913
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
+ QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W++++K+
Sbjct: 914 YITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLKQSGANWYETIKS 970
Query: 1004 ALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L+ G + + +F K+ + ++VDD+I+ N + I L + K
Sbjct: 971 YLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
Query: 1063 --DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT------G 1112
+LG E+Q D G+ +K + KY+ K MEN+ P + G
Sbjct: 1027 IINLGESDN----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVPLNPKG 1077
Query: 1113 RHFTVEGE-----------------KLKDPTVFRQAIGGLQYLTHT-RPDIAFSVNKLSQ 1154
R + G+ K+K + ++ IG Y+ + R D+ + +N L+Q
Sbjct: 1078 RKLSAPGQPGLYIDQQELELEEDDYKMKVHEM-QKLIGLASYVGYKFRFDLLYYINTLAQ 1136
Query: 1155 YMSSPTTDHWQGIKRILRYLQGTINYCL------HIKPSTDLDITGFSDADWATSIDDRK 1208
++ P+ +++++ T + L +KP+ L + SDA + K
Sbjct: 1137 HILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVV--ISDASYGNQ-PYYK 1193
Query: 1209 SMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRK 1268
S G L +I S K + S+TE+E A+ + + + L+ EL K
Sbjct: 1194 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITK 1253
Query: 1269 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKP 1328
+L + + SN R++ +RD+V N + V Y+ T IAD +TKP
Sbjct: 1254 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1313
Query: 1329 LSHTRFSQLRDK 1340
L F L +K
Sbjct: 1314 LPIKTFKLLTNK 1325
Score = 98.2 bits (243), Expect = 1e-19
Identities = 82/310 (26%), Positives = 133/310 (42%), Gaps = 57/310 (18%)
Query: 366 VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKL--TGKVILKGLLK--------- 414
VL+ PNI +LLS+++LAA D CF K L + +L ++K
Sbjct: 89 VLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIVKYGDFYWVSK 141
Query: 415 -------------NGLYQLSDTKGNPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVP 461
N ++ T+ PY F+ HR L H N + + LK+ +
Sbjct: 142 KYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRYSLKNNTITYF 195
Query: 462 PSDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSS 510
+ + C C GK H+ + ++ EP + +HTD++GP + S
Sbjct: 196 NESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKS 255
Query: 511 GFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKVIQCDGGGEY- 562
Y++ F D+ ++F W+YPL + E + FTT +NQF + VIQ D G EY
Sbjct: 256 APSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYT 315
Query: 563 -KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFST 621
+ + K GI + + +G AER +R + + T L + +P H W+ A
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 622 AVYLINRLPS 631
+ + N L S
Sbjct: 376 STIVRNSLAS 385
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 122 bits (307), Expect = 5e-27
Identities = 163/672 (24%), Positives = 290/672 (42%), Gaps = 92/672 (13%)
Query: 730 INNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN-----GSSEG 784
I P+T + + I D +P + E+ E + + ++ QT++ G S
Sbjct: 1112 IKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELPPINSRQTNSSLGGIGDSNA 1171
Query: 785 DTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITETYKDTV 839
TT + + + + + S DT NT N RSK IH + + ++ K
Sbjct: 1172 YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKSIKPIR 1229
Query: 840 EPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY 892
EA+T + + EA KE L+ KTW Y D++ I D K V + +
Sbjct: 1230 TTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEI-DPKRVINSMF 1288
Query: 893 ----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNW 943
K DG+ KAR VA+G I + +T+ ++ +TV LS+A+ N+
Sbjct: 1289 IFNKKRDGT---HKARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNY 1340
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
+ QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W++++K+
Sbjct: 1341 YITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLKQSGANWYETIKS 1397
Query: 1004 ALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L+ G + + +F K+ + ++VDD+++ + N + I++L + K
Sbjct: 1398 YLIQQCGMEEVRGWSCVF----KNSQVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTK 1453
Query: 1063 --DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT------G 1112
+LG E+Q D G+ +K + KY+ K MEN+ P + G
Sbjct: 1454 IINLGESDE----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVPLNPKG 1504
Query: 1113 RHFTVEGE-----------------KLKDPTVFRQAIGGLQYLTHT-RPDIAFSVNKLSQ 1154
R + G+ K+K + ++ IG Y+ + R D+ + +N L+Q
Sbjct: 1505 RKLSAPGQPGLYIDQQELELEEDDYKMKVHEM-QKLIGLASYVGYKFRFDLLYYINTLAQ 1563
Query: 1155 YMSSPTTDHWQGIKRILRYLQGTINYCL------HIKPSTDLDITGFSDADWATSIDDRK 1208
++ P+ +++++ T + L +KP+ L + SDA + K
Sbjct: 1564 HILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVV--ISDASYGNQ-PYYK 1620
Query: 1209 SMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRK 1268
S G L +I S K + S+TE+E A+ + + + L+ EL K
Sbjct: 1621 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITK 1680
Query: 1269 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKP 1328
+L + + SN R++ +RD+V N + V Y+ T IAD +TKP
Sbjct: 1681 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1740
Query: 1329 LSHTRFSQLRDK 1340
L F L +K
Sbjct: 1741 LPIKTFKLLTNK 1752
Score = 100 bits (248), Expect = 4e-20
Identities = 143/680 (21%), Positives = 252/680 (37%), Gaps = 117/680 (17%)
Query: 23 NNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQRLLGW 82
N++P W + ++ L G + P IT + + N F+ + A Q L W
Sbjct: 179 NDFPNWVKTYIKFLQNSNLGGIIPTVNGKPVRQITDDELTFLYN-TFQIF-APSQFLPTW 236
Query: 83 MLNSMATEMATQL-LHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKMEDYL 141
+ + ++ + + + ++ +++ + Q T + + Y G + +
Sbjct: 237 VKDILSVDYTDIMKILSKSIEKMQSDTQEANDIVTLANLQY---------NGSTPADAFE 287
Query: 142 IKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLTFE 201
K+ N+ D+L G I+N + GL EY + H ++ +L L
Sbjct: 288 TKVTNIIDRLNNNGIHINNKVACQLIMRGLSGEYKFLRYTRHRHLNMTVAEL---FLDIH 344
Query: 202 SRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQ 261
+ E+ N N N ++ + D+R +N K +
Sbjct: 345 AIYEEQQGSRNSKPNYRRNPSD--EKNDSRSYTNTT-----------------KPKVIAR 385
Query: 262 VYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVEDY--DWYFDSGASN 319
KTN++ + + S ++ S D+D N+ D N
Sbjct: 386 NPQKTNNSKSKTARAHNVSTSNNSPSTDNDSISKSTTEPIQLNNKHDLILGQKLTESTVN 445
Query: 320 HVTHQTDKFQD-------------LTEHH----GKNSQV-VGNGDKLEIVATGSSKLKSL 361
H H D+ + HH N + V + K I L+
Sbjct: 446 HTNHSDDELPGHLLLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFH 505
Query: 362 NLDD------VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKL--TGKVILKGLL 413
D+ VL+ PNI +LLS+++LAA D CF K L + +L ++
Sbjct: 506 FQDNTKTSIKVLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIV 558
Query: 414 K----------------------NGLYQLSDTKGNPYAFVSVKESWHRRLGHPNNKVLDK 451
K N ++ T+ PY F+ HR L H N + +
Sbjct: 559 KYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRY 612
Query: 452 VLKSCNVKVPPSDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDV 500
LK+ + + + C C GK H+ + ++ EP + +HTD+
Sbjct: 613 SLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDI 672
Query: 501 WGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKV 553
+GP + S Y++ F D+ ++F W+YPL + E + FTT +NQF + V
Sbjct: 673 FGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLV 732
Query: 554 IQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMP 611
IQ D G EY + + K GI + + +G AER +R + + T L + +P
Sbjct: 733 IQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLP 792
Query: 612 LHYWWEAFSTAVYLINRLPS 631
H W+ A + + N L S
Sbjct: 793 NHLWFSAIEFSTIVRNSLAS 812
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 122 bits (306), Expect = 7e-27
Identities = 69/128 (53%), Positives = 87/128 (67%), Gaps = 3/128 (2%)
Query: 815 TRSKSGIHKPKLPY-IGITETYKDTVEPANVKEALTRTLWKEAMQKEFQALMSNKTWILV 873
TRSK+GI+K Y + IT T K EP +V AL W +AMQ+E AL NKTWILV
Sbjct: 3 TRSKAGINKLNPKYSLTITTTIKK--EPKSVIFALKDPGWCQAMQEELDALSRNKTWILV 60
Query: 874 PYQDQENIVDSKWVFKTKYKSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVRV 933
P +NI+ KWVFKTK SDG+++R KARLVAKGF Q GI + ET+SPVV+ +T+R
Sbjct: 61 PPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRT 120
Query: 934 ILSIAVHL 941
IL++A L
Sbjct: 121 ILNVAQQL 128
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 120 bits (302), Expect = 2e-26
Identities = 163/677 (24%), Positives = 292/677 (43%), Gaps = 96/677 (14%)
Query: 725 PLKTTINNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN----- 779
P+KT+ P + + + I D +P + E E ++ + ++ QT++
Sbjct: 684 PIKTSDTCPKEN----TEESIIADLPLPDLPPEPPTELSDSFKELPPINSRQTNSSLGGI 739
Query: 780 GSSEGDTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITET 834
G S TT + + + + + S DT NT N RSK IH + + ++
Sbjct: 740 GDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKS 797
Query: 835 YKDTVEPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWV 887
K EA+T + + EA KE L+ KTW Y D++ I D K V
Sbjct: 798 IKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEI-DPKRV 856
Query: 888 FKTKY----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIA 938
+ + K DG+ KAR VA+G I + +T+ ++ +TV LS+A
Sbjct: 857 INSMFIFNRKRDGT---HKARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLA 908
Query: 939 VHLNWEVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWF 998
+ N+ + QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W+
Sbjct: 909 LDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLKQSGANWY 965
Query: 999 DSLKTALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLND 1057
+++K+ L+ G + + +F ++ + ++VDD+++ + N + I +L
Sbjct: 966 ETIKSYLIKQCGMEEVRGWSCVF----ENSQVTICLFVDDMVLFSKNLNSNKRIIDKLKM 1021
Query: 1058 VFSLK--DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT-- 1111
+ K +LG E+Q D G+ +K + KY+ K MEN+ P +
Sbjct: 1022 QYDTKIINLGESDE----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVP 1072
Query: 1112 ----GRHFTVEGE-----------------KLKDPTVFRQAIGGLQYLTHT-RPDIAFSV 1149
GR + G+ K+K + ++ IG Y+ + R D+ + +
Sbjct: 1073 LNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEM-QKLIGLASYVGYKFRFDLLYYI 1131
Query: 1150 NKLSQYMSSPTTDHWQGIKRILRYLQGTINYCL------HIKPSTDLDITGFSDADWATS 1203
N L+Q++ P+ +++++ T + L +KP+ L + SDA +
Sbjct: 1132 NTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVV--ISDASYGNQ 1189
Query: 1204 IDDRKSMAGQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKL 1263
KS G L +I S K + S+TE+E A+ + + + L+ EL
Sbjct: 1190 -PYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDK 1248
Query: 1264 PLPRKPILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIAD 1323
K +L + + SN R++ +RD+V N + V Y+ T IAD
Sbjct: 1249 KPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIAD 1308
Query: 1324 CLTKPLSHTRFSQLRDK 1340
+TKPL F L +K
Sbjct: 1309 VMTKPLPIKTFKLLTNK 1325
Score = 98.2 bits (243), Expect = 1e-19
Identities = 82/310 (26%), Positives = 133/310 (42%), Gaps = 57/310 (18%)
Query: 366 VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKL--TGKVILKGLLK--------- 414
VL+ PNI +LLS+++LAA D CF K L + +L ++K
Sbjct: 89 VLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIVKYGDFYWVSK 141
Query: 415 -------------NGLYQLSDTKGNPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVP 461
N ++ T+ PY F+ HR L H N + + LK+ +
Sbjct: 142 KYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRYSLKNNTITYF 195
Query: 462 PSDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSS 510
+ + C C GK H+ + ++ EP + +HTD++GP + S
Sbjct: 196 NESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKS 255
Query: 511 GFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKVIQCDGGGEY- 562
Y++ F D+ ++F W+YPL + E + FTT +NQF + VIQ D G EY
Sbjct: 256 APSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYT 315
Query: 563 -KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFST 621
+ + K GI + + +G AER +R + + T L + +P H W+ A
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 622 AVYLINRLPS 631
+ + N L S
Sbjct: 376 STIVRNSLAS 385
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 120 bits (302), Expect = 2e-26
Identities = 163/669 (24%), Positives = 284/669 (42%), Gaps = 86/669 (12%)
Query: 730 INNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN-----GSSEG 784
I P+T + + I D +P + E+ E + + ++ QT++ G S
Sbjct: 685 IKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELPPINSHQTNSSLGGIGDSNA 744
Query: 785 DTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITETYKDTV 839
TT + + + + + S DT NT N RSK IH + + ++ K
Sbjct: 745 YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKSIKPIR 802
Query: 840 EPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY 892
EA+T + + EA KE L+ TW Y D++ I D K V + +
Sbjct: 803 TTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEI-DPKRVINSMF 861
Query: 893 ----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNW 943
K DG+ KAR VA+G I + +T+ ++ +TV LS+A+ N+
Sbjct: 862 IFNRKRDGT---HKARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNY 913
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
+ QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W++++K+
Sbjct: 914 YITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLKQSGANWYETIKS 970
Query: 1004 ALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L+ G + + +F K+ + ++VDD+I+ N + I L + K
Sbjct: 971 YLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
Query: 1063 --DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT------G 1112
+LG E+Q D G+ +K + KY+ K MEN+ P + G
Sbjct: 1027 IINLGESDN----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVPLNPKG 1077
Query: 1113 RHFTVEGE---------------KLKDPTVFRQAIGGL-QYLTHT-RPDIAFSVNKLSQY 1155
R + G+ + K+ Q + GL Y+ + R D+ + +N L+Q+
Sbjct: 1078 RKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQH 1137
Query: 1156 MSSPTTDHWQGIKRILRYLQGTINYCL--HIKPSTDLD--ITGFSDADWATSIDDRKSMA 1211
+ P+ +++++ T + L H T+ D + SDA + KS
Sbjct: 1138 ILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQ-PYYKSQI 1196
Query: 1212 GQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPIL 1271
G L +I S K + S+TE+E A+ + + + L+ EL K +L
Sbjct: 1197 GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITKGLL 1256
Query: 1272 WCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSH 1331
+ + SN R++ +RD+V N + V Y+ T IAD +TKPL
Sbjct: 1257 TDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPI 1316
Query: 1332 TRFSQLRDK 1340
F L +K
Sbjct: 1317 KTFKLLTNK 1325
Score = 98.2 bits (243), Expect = 1e-19
Identities = 82/310 (26%), Positives = 133/310 (42%), Gaps = 57/310 (18%)
Query: 366 VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKL--TGKVILKGLLK--------- 414
VL+ PNI +LLS+++LAA D CF K L + +L ++K
Sbjct: 89 VLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIVKYGDFYWVSK 141
Query: 415 -------------NGLYQLSDTKGNPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVKVP 461
N ++ T+ PY F+ HR L H N + + LK+ +
Sbjct: 142 KYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRYSLKNNTITYF 195
Query: 462 PSDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSS 510
+ + C C GK H+ + ++ EP + +HTD++GP + S
Sbjct: 196 NESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKS 255
Query: 511 GFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKVIQCDGGGEY- 562
Y++ F D+ ++F W+YPL + E + FTT +NQF + VIQ D G EY
Sbjct: 256 APSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYT 315
Query: 563 -KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFST 621
+ + K GI + + +G AER +R + + T L + +P H W+ A
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 622 AVYLINRLPS 631
+ + N L S
Sbjct: 376 STIVRNSLAS 385
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 120 bits (300), Expect = 3e-26
Identities = 163/669 (24%), Positives = 284/669 (42%), Gaps = 86/669 (12%)
Query: 730 INNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN-----GSSEG 784
I P+T + + I D +P + E+ E + + ++ QT++ G S
Sbjct: 685 IKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELPPINSHQTNSSLGGIGDSNA 744
Query: 785 DTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITETYKDTV 839
TT + + + + + S DT NT N RSK IH + + ++ K
Sbjct: 745 YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKSIKPIR 802
Query: 840 EPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY 892
EA+T + + EA KE L+ KTW Y D++ I D K V + +
Sbjct: 803 TTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEI-DPKRVINSMF 861
Query: 893 ----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNW 943
K DG+ KAR VA+G I + +T+ ++ +TV LS+A+ N+
Sbjct: 862 IFNKKRDGT---HKARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNY 913
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
+ QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W++++K+
Sbjct: 914 YITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLKQSGANWYETIKS 970
Query: 1004 ALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L+ G + + +F K+ + ++VDD+I+ N + I L + K
Sbjct: 971 YLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
Query: 1063 --DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT------G 1112
+LG E+Q D G+ +K + KY+ K MEN+ P + G
Sbjct: 1027 IINLGESDN----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVPLNPKG 1077
Query: 1113 RHFTVEGE---------------KLKDPTVFRQAIGGL-QYLTHT-RPDIAFSVNKLSQY 1155
R + G+ + K+ Q + GL Y+ + R D+ + +N L+Q+
Sbjct: 1078 RKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQH 1137
Query: 1156 MSSPTTDHWQGIKRILRYLQGTINYCL--HIKPSTDLD--ITGFSDADWATSIDDRKSMA 1211
+ P+ +++++ T + L H T+ D + SDA + KS
Sbjct: 1138 ILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQ-PYYKSQI 1196
Query: 1212 GQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPIL 1271
G L +I S K + S+TE+E A+ + + + L+ EL K +L
Sbjct: 1197 GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPIIKGLL 1256
Query: 1272 WCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSH 1331
+ + S R++ +RD+V N + V Y+ T IAD +TKPL
Sbjct: 1257 TDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPI 1316
Query: 1332 TRFSQLRDK 1340
F L +K
Sbjct: 1317 KTFKLLTNK 1325
Score = 98.6 bits (244), Expect = 1e-19
Identities = 84/310 (27%), Positives = 134/310 (43%), Gaps = 57/310 (18%)
Query: 366 VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKL--TGKVILKGLLK--------- 414
VL+ PNI +LLS+++LAA D CF K L + +L ++K
Sbjct: 89 VLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIVKYGDFYWVSK 141
Query: 415 -------------NGLYQLSDTKGNPYAFVSVKESWHRRLGHPNNKVLDKVLKSCNVK-- 459
N ++ T+ PY F+ HR L H N + + LK+ +
Sbjct: 142 KYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRYSLKNNTITYF 195
Query: 460 ----VPPSDNFSF-CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSS 510
V S + C C GK H+ + ++ EP + +HTD++GP + S
Sbjct: 196 NESDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKS 255
Query: 511 GFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKVIQCDGGGEY- 562
Y++ F D+ ++F W+YPL + E + FTT +NQF + VIQ D G EY
Sbjct: 256 APSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYT 315
Query: 563 -KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFST 621
+ + K GI + + +G AER +R + + T L + +P H W+ A
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 622 AVYLINRLPS 631
+ + N L S
Sbjct: 376 STIVRNSLAS 385
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 118 bits (296), Expect = 1e-25
Identities = 158/657 (24%), Positives = 279/657 (42%), Gaps = 68/657 (10%)
Query: 733 PSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDNGSSEGD------T 786
P S +A + ++D +P + ++ +T++ + QT++ D T
Sbjct: 1130 PDKSSKNVTADSILDDLPLPDLTHQSPTDTSDVSKDIPHIHSRQTNSSLGGMDDSNVLTT 1189
Query: 787 THEETLDIVQQQNVGESSLDT----NTSNAIHTRSKSGIHKPKLPYIGITETYKDTVEPA 842
T + + + E S DT N + RSK I+ + I ++ K
Sbjct: 1190 TKSKKRSLEDNETEIEVSRDTWNNKNMRSLEPPRSKKRINL--IAAIKGVKSIKPVRTTL 1247
Query: 843 NVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY--- 892
EA+T + + EA KE L+ TW Y D+ N +D K V + +
Sbjct: 1248 RYDEAITYNKDNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDR-NDIDPKKVINSMFIFN 1306
Query: 893 -KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNWEVR 946
K DG+ KAR VA+G I + +T+ ++ +TV LSIA+ ++ +
Sbjct: 1307 KKRDGT---HKARFVARG-----DIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYIT 1358
Query: 947 QLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKTALL 1006
QLDI++A+L +KE +++ P K + +L K++YGLKQ+ W++++K+ L+
Sbjct: 1359 QLDISSAYLYADIKEELYIRPPPHLGLNDK---LLRLRKSLYGLKQSGANWYETIKSYLI 1415
Query: 1007 NW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK--D 1063
N Q + +F K+ + ++VDD+I+ N + I L + K +
Sbjct: 1416 NCCDMQEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIIN 1471
Query: 1064 LGR----LHY-FLGIEVQRDASG-MYLKQSKYIGDLLKKF--------KMENASPCPTPM 1109
LG + Y LG+E++ S M L K + + L K K A P
Sbjct: 1472 LGESDNEIQYDILGLEIKYQRSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHY 1531
Query: 1110 ITGRHFTVEGEKLKDPTVFRQAIGGL-QYLTHT-RPDIAFSVNKLSQYMSSPTTDHWQGI 1167
I ++ ++ K+ Q + GL Y+ + R D+ + +N L+Q++ P+
Sbjct: 1532 IDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMT 1591
Query: 1168 KRILRYLQGTINYCL---HIKPST-DLDITGFSDADWATSIDDRKSMAGQCVFLGETLIS 1223
+++++ T + L KP+ D + SDA + KS G L +I
Sbjct: 1592 YELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASYGNQ-PYYKSQIGNIFLLNGKVIG 1650
Query: 1224 WSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPILWCDNLSAKALAS 1283
S K + S+TE+E A+ + + + L+ EL K +L + + S
Sbjct: 1651 GKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKS 1710
Query: 1284 NPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSHTRFSQLRDK 1340
R++ +RD+V N + V Y+ T IAD +TKPL F L +K
Sbjct: 1711 TNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
Score = 113 bits (283), Expect = 3e-24
Identities = 138/549 (25%), Positives = 227/549 (41%), Gaps = 65/549 (11%)
Query: 133 GEMKMEDYLIKMKNLADKLKLAGNPISNS---NLIIQTLNGLDSEYNPIVVKLSDHTTLS 189
G + + I + + +LK +S+ LI++ L+G D +Y + + LS
Sbjct: 275 GSTSADTFEITVSTIIQRLKENNINVSDRLACQLILKGLSG-DFKYLRNQYRTKTNMKLS 333
Query: 190 WVDLQAQLLTFESRIEQLNTLTNLNLNAT-ANVANKFDHRDNRFNSNNNWRGSNFRGWRG 248
+ + QL+ E++I LN + ++ NV+ + N + N++ +N
Sbjct: 334 QLFAEIQLIYDENKIMNLNKPSQYKQHSEYKNVSRTSPNTTNTKVTTRNYQRTN------ 387
Query: 249 GRGRGRSSKAPCQVYGKTNHTAINCFHRFDKNYSRSNYSADSD-------KQGSHNAFIA 301
+ R++KA + + + +N H + S S D++ K+ I
Sbjct: 388 -SSKPRAAKAH-NIATSSKFSRVNNDHINESTVSSQYLSDDNELSLGQQQKESKPTHTID 445
Query: 302 SQNSVEDYDWYFDSGASNHVTHQTDKFQDLTEHHGKNSQVVGNGDKLEIVATGSSKLKSL 361
S + + D+ DSGAS + T + N V + I A G+
Sbjct: 446 SNDELPDH-LLIDSGASQTLVRSAHYLHHATPNSEINI-VDAQKQDIPINAIGNLHFNFQ 503
Query: 362 NLDDV----LYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVKEKLTGKVILKGLLKNGL 417
N L+ PNI +LLS+S+LA + NI F +N E+ G V+ +
Sbjct: 504 NGTKTSIKALHTPNIAYDLLSLSELA-NQNITACFTRNTL---ERSDGTVLAPIVKHGDF 559
Query: 418 YQLSD-----------TKGNPYAFVSVKES----WHRRLGHPNNKVLDKVLKSCNVKVPP 462
Y LS T N SV + HR LGH N + + K LK V
Sbjct: 560 YWLSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLK 619
Query: 463 SDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDVWGPAPIMSSSG 511
+ + C C GK H+ + + EP + +HTD++GP + S
Sbjct: 620 ESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSA 679
Query: 512 FKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKVIQCDGGGEY-- 562
Y++ F D+ +RF W+YPL + E + FT+ +NQFN R+ VIQ D G EY
Sbjct: 680 PSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTN 739
Query: 563 KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTA 622
K + K + GI + + +G AER +R + TLL + +P H W+ A +
Sbjct: 740 KTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFS 799
Query: 623 VYLINRLPS 631
+ N L S
Sbjct: 800 TIIRNSLVS 808
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 118 bits (295), Expect = 1e-25
Identities = 163/669 (24%), Positives = 283/669 (41%), Gaps = 86/669 (12%)
Query: 730 INNPSTSFPLCSAGNSINDASIPIIEEENQDETNEEDSQGVTSDTEQTDN-----GSSEG 784
I P+T + + I D +P + E+ E + + ++ QT++ G S
Sbjct: 1112 IKTPTTVSEQNTEESIIADLPLPDLPPESPTEFPDPFKELPPINSHQTNSSLGGIGDSNA 1171
Query: 785 DTT-HEETLDIVQQQNVGESSLDT-NTSNAIHT---RSKSGIHKPKLPYIGITETYKDTV 839
TT + + + + + S DT NT N RSK IH + + ++ K
Sbjct: 1172 YTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIHL--IAAVKAVKSIKPIR 1229
Query: 840 EPANVKEALT-------RTLWKEAMQKEFQALMSNKTWILVPYQDQENIVDSKWVFKTKY 892
EA+T + + EA KE L+ KTW Y D++ I D K V + +
Sbjct: 1230 TTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEI-DPKRVINSMF 1288
Query: 893 ----KSDGSIERRKARLVAKGFQQTAGIDYEETFSPVVKVSTVR-----VILSIAVHLNW 943
K DG+ KAR VA+G I + +T+ ++ +TV LS+A+ N+
Sbjct: 1289 IFNKKRDGT---HKARFVARG-----DIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNY 1340
Query: 944 EVRQLDINNAFLNGYLKETVFMHQPEGFVDPTKPNHICKLSKAIYGLKQAPRAWFDSLKT 1003
+ QLDI++A+L +KE +++ P K + +L K+ YGLKQ+ W++++K+
Sbjct: 1341 YITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSHYGLKQSGANWYETIKS 1397
Query: 1004 ALLNW-GFQNTKSDPSLFLLKGKDHITFLLIYVDDIIVTGSSNNFLQAFIKQLNDVFSLK 1062
L+ G + + +F K+ + ++VDD+I+ N + I L + K
Sbjct: 1398 YLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1453
Query: 1063 --DLGRLHYFLGIEVQRDASGMYLK--QSKYIGDLLKKFKMENASPCPTPMIT------G 1112
+LG E+Q D G+ +K + KY+ K MEN+ P + G
Sbjct: 1454 IINLGESDN----EIQYDILGLEIKYQRGKYM-----KLGMENSLTEKIPKLNVPLNPKG 1504
Query: 1113 RHFTVEGE---------------KLKDPTVFRQAIGGL-QYLTHT-RPDIAFSVNKLSQY 1155
R + G+ + K+ Q + GL Y+ + R D+ + +N L+Q+
Sbjct: 1505 RKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQH 1564
Query: 1156 MSSPTTDHWQGIKRILRYLQGTINYCL--HIKPSTDLD--ITGFSDADWATSIDDRKSMA 1211
+ P+ +++++ T + L H T+ D + SDA + KS
Sbjct: 1565 ILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQ-PYYKSQI 1623
Query: 1212 GQCVFLGETLISWSSRKQKVVSRSSTESEYRALVDLAAEIAWIHSLLFELKLPLPRKPIL 1271
G L +I S K + S+TE+E A+ + + + L+ EL K +L
Sbjct: 1624 GNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPIIKGLL 1683
Query: 1272 WCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNKVVVAYVPTTDQIADCLTKPLSH 1331
+ + S R++ +RD+V N + V Y+ T IAD +TKPL
Sbjct: 1684 TDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPI 1743
Query: 1332 TRFSQLRDK 1340
F L +K
Sbjct: 1744 KTFKLLTNK 1752
Score = 100 bits (248), Expect = 4e-20
Identities = 145/680 (21%), Positives = 254/680 (37%), Gaps = 117/680 (17%)
Query: 23 NNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQRLLGW 82
N++P W + ++ L G + P IT + + N F+ + A Q L W
Sbjct: 179 NDFPNWVKTYIKFLQNSNLGGIIPTVNGKPVRQITDDELTFLYN-TFQIF-APSQFLPTW 236
Query: 83 MLNSMATEMATQL-LHCETSKQLWDEAQSLAGAHTRSQIIYLKFQFHSIRKGEMKMEDYL 141
+ + ++ + + + ++ +++ + Q T + + Y G + +
Sbjct: 237 VKDILSVDYTDIMKILSKSIEKMQSDTQEANDIVTLANLQY---------NGSTPADAFE 287
Query: 142 IKMKNLADKLKLAGNPISNSNLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLTFE 201
K+ N+ D+L G I+N + GL EY + H ++ +L L
Sbjct: 288 TKVTNIIDRLNNNGIHINNKVACQLIMRGLSGEYKFLRYTRHRHLNMTVAEL---FLDIH 344
Query: 202 SRIEQLNTLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQ 261
+ E+ N N N+++ + D+R +N K +
Sbjct: 345 AIYEEQQGSRNSKPNYRRNLSD--EKNDSRSYTNTT-----------------KPKVIAR 385
Query: 262 VYGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVEDYDWYFDSGAS--N 319
KTN++ + + S ++ S D+D N+ D + S N
Sbjct: 386 NPQKTNNSKSKTARAHNVSTSNNSPSTDNDSISKSTTEPIQLNNKHDLTLGQELTESTVN 445
Query: 320 HVTHQTDKFQD-------------LTEHH----GKNSQV-VGNGDKLEIVATGSSKLKSL 361
H H D+ + HH N + V + K I L+
Sbjct: 446 HTNHSDDELPGHLLLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFH 505
Query: 362 NLDD------VLYVPNITKNLLSVSKLAADNNIFVEFDKNCCFVK---EKLTGKVILKGL 412
D+ VL+ PNI +LLS+++LAA D CF K E+ G V+ +
Sbjct: 506 FQDNTKTSIKVLHTPNIAYDLLSLNELAA-------VDITACFTKNVLERSDGTVLAPIV 558
Query: 413 LKNGLYQLS---------------------DTKGNPYAFVSVKESWHRRLGHPNNKVLDK 451
Y +S T+ PY F+ HR L H N + +
Sbjct: 559 QYGDFYWVSKRYLLPSNISVPTINNVHTSESTRKYPYPFI------HRMLAHANAQTIRY 612
Query: 452 VLKSCNVKVPPSDNFSF-------CEACQYGKM----HLLPFKSSSSHAQEPLELVHTDV 500
LK+ + + + C C GK H+ + ++ EP + +HTD+
Sbjct: 613 SLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDI 672
Query: 501 WGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSE--TVQAFTT-----ENQFNKRIKV 553
+GP + S Y++ F D+ ++F W+YPL + E + FTT +NQF + V
Sbjct: 673 FGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLV 732
Query: 554 IQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMP 611
IQ D G EY + + K GI + + +G AER +R + + T L + +P
Sbjct: 733 IQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLP 792
Query: 612 LHYWWEAFSTAVYLINRLPS 631
H W+ A + + N L S
Sbjct: 793 NHLWFSAIEFSTIVRNSLAS 812
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 76.3 bits (186), Expect = 5e-13
Identities = 33/78 (42%), Positives = 52/78 (66%)
Query: 1137 YLTHTRPDIAFSVNKLSQYMSSPTTDHWQGIKRILRYLQGTINYCLHIKPSTDLDITGFS 1196
YLT TRPD+ F+VN+LSQ+ S+ T Q + ++L Y++GT+ L ++DL + F+
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 1197 DADWATSIDDRKSMAGQC 1214
D+DWA+ D R+S+ G C
Sbjct: 62 DSDWASCPDTRRSVTGFC 79
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 57.8 bits (138), Expect = 2e-07
Identities = 32/102 (31%), Positives = 52/102 (50%), Gaps = 5/102 (4%)
Query: 406 KVILKGLLKNGLYQLS---DTKGNPYAFVSVKES--WHRRLGHPNNKVLDKVLKSCNVKV 460
+ ILKG + LY L +T + A + E+ WH RL H + + ++ ++K +
Sbjct: 36 RTILKGNRHDSLYILQGSVETGESNLAETAKDETRLWHSRLAHMSQRGMELLVKKGFLDS 95
Query: 461 PPSDNFSFCEACQYGKMHLLPFKSSSSHAQEPLELVHTDVWG 502
+ FCE C YGK H + F + + PL+ VH+D+WG
Sbjct: 96 SKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWG 137
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 52.4 bits (124), Expect = 8e-06
Identities = 44/163 (26%), Positives = 67/163 (40%), Gaps = 9/163 (5%)
Query: 490 QEPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFTTENQFNK 549
Q+P + D GP P S G+ Y + +D + FTW+YP K S + + +
Sbjct: 673 QKPFDKFFIDYIGPLP--PSQGYLYVLVVVDGMTGFTWLYPTKAPSTSATVKSLNVLTSI 730
Query: 550 RI-KVIQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLA 606
I KVI D G + + A + GI S PY Q + ERK+ I LL
Sbjct: 731 AIPKVIHSDQGAAFTSSTFAEWAKERGIHLEFSTPYHPQSGSKVERKNSDIKRLLTKLLV 790
Query: 607 QAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPN 649
W++ +N S V + +P+ L+F + N
Sbjct: 791 GRPTK---WYDLLPVVQLALNNTYSPVLK-YTPHQLLFGIDSN 829
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 51.6 bits (122), Expect = 1e-05
Identities = 42/144 (29%), Positives = 69/144 (47%), Gaps = 9/144 (6%)
Query: 510 SGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFTTENQFNKRI---KVIQCDGGGEY--KP 564
+G+KY + F+D FS + YP +Q++ + A + R KVI D G + +
Sbjct: 777 AGYKYLLVFVDTFSGWVEAYPTRQETAHMVAKKILEEIFPRFGLPKVIGSDNGPAFVSQV 836
Query: 565 VQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVY 624
Q LA +GI +++ C Y Q +G+ ER +R I E LT L + L W S A+
Sbjct: 837 SQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKE-TLTKLT-LETGLKDWRRLLSLALL 894
Query: 625 LINRLPSQVTQNESPYSLIFHKEP 648
P++ +PY +++ P
Sbjct: 895 RARNTPNRF--GLTPYEILYGGPP 916
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 50.4 bits (119), Expect = 3e-05
Identities = 43/163 (26%), Positives = 68/163 (41%), Gaps = 11/163 (6%)
Query: 491 EPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLK--QKSETVQAFTTENQFN 548
+P + + D GP P S+G+ + + +D + F W+YP K S TV+A
Sbjct: 882 KPFDKFYIDYIGPLP--PSNGYLHVLVVVDSMTGFVWLYPTKAPSTSATVKALNMLTSI- 938
Query: 549 KRIKVIQCDGGGEY--KPVQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLA 606
KV+ D G + A + GIQ S PY Q +G+ ERK+ I LL
Sbjct: 939 AIPKVLHSDQGAAFTSSTFADWAKEKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLI 998
Query: 607 QAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPN 649
W++ +N S + +P+ L+F + N
Sbjct: 999 GRPAK---WYDLLPVVQLALNNSYSP-SSKYTPHQLLFGVDSN 1037
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 50.1 bits (118), Expect = 4e-05
Identities = 44/163 (26%), Positives = 67/163 (40%), Gaps = 11/163 (6%)
Query: 491 EPLELVHTDVWGPAPIMSSSGFKYYVHFIDDFSRFTWIYPLK--QKSETVQAFTTENQFN 548
+P + D GP P S+G+ + + +D + F W+YP K S TV+A
Sbjct: 884 KPFDKFFIDYIGPLP--PSNGYLHVLVVVDSMTGFVWLYPTKAPSTSATVKALNMLTSIA 941
Query: 549 KRIKVIQCDGGGEYKPV--QKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLA 606
KVI D G + A + GIQ S PY Q +G+ ERK+ I LL
Sbjct: 942 VP-KVIHSDQGAAFTSATFADWAKNKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLV 1000
Query: 607 QAQMPLHYWWEAFSTAVYLINRLPSQVTQNESPYSLIFHKEPN 649
W++ +N S + +P+ L+F + N
Sbjct: 1001 GRPAK---WYDLLPVVQLALNNSYSP-SSKYTPHQLLFGIDSN 1039
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 50.1 bits (118), Expect = 4e-05
Identities = 41/144 (28%), Positives = 69/144 (47%), Gaps = 9/144 (6%)
Query: 510 SGFKYYVHFIDDFSRFTWIYPLKQKSETVQAFTTENQFNKRI---KVIQCDGGGEY--KP 564
+G+KY + F+D FS + +P +Q++ + A + R KVI D G + +
Sbjct: 920 AGYKYLLVFVDTFSGWVEAFPTRQETAHIVAKKILEEIFPRFGLPKVIGSDNGPAFVSQV 979
Query: 565 VQKLAIDVGIQFRMSCPYTFQQNGRAERKHRHIAEFGLTLLAQAQMPLHYWWEAFSTAVY 624
Q LA +GI +++ C Y Q +G+ ER +R I E LT L + L W S A+
Sbjct: 980 SQGLARILGINWKLHCAYRPQSSGQVERMNRTIKE-TLTKLT-LETGLKDWRRLLSLALL 1037
Query: 625 LINRLPSQVTQNESPYSLIFHKEP 648
P++ +PY +++ P
Sbjct: 1038 RARNTPNRF--GLTPYEILYGGPP 1059
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 160,612,032
Number of Sequences: 164201
Number of extensions: 6929030
Number of successful extensions: 19604
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 19223
Number of HSP's gapped (non-prelim): 289
length of query: 1349
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1227
effective length of database: 39,941,532
effective search space: 49008259764
effective search space used: 49008259764
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146790.9


