

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.3 + phase: 0 /pseudo
(1363 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
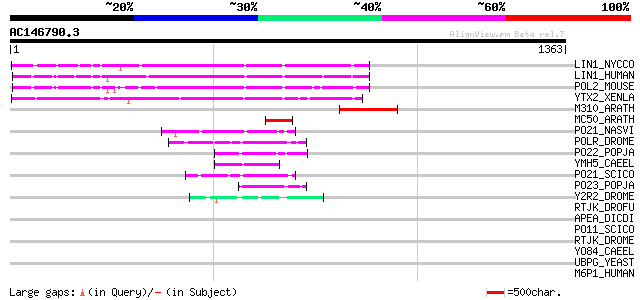
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 181 1e-44
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 176 3e-43
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 172 6e-42
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 159 6e-38
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 129 5e-29
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 91 3e-17
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 80 5e-14
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 74 2e-12
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 74 3e-12
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 65 2e-09
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 59 7e-08
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 58 2e-07
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 56 8e-07
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 43 0.005
APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (E... 43 0.005
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 42 0.011
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 41 0.019
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 36 0.63
UBPG_YEAST (Q02863) Ubiquitin carboxyl-terminal hydrolase 16 (EC... 35 1.4
M6P1_HUMAN (O60664) Mannose-6-phosphate receptor binding protein... 34 2.4
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 181 bits (459), Expect = 1e-44
Identities = 206/911 (22%), Positives = 394/911 (42%), Gaps = 62/911 (6%)
Query: 4 MKLLSWNCRGLGNPRAIRALLRLIRIQNPQVVFLMETRLKSDEMERVRINCGFSSGLFVP 63
+ + S N GL P L I+ P + + E+ L + R+++ G+SS +F
Sbjct: 7 LSIFSINVNGLNCPLKRHRLADWIQKLKPDICCIQESHLTLKDKYRLKVK-GWSS-IF-- 62
Query: 64 CDGFGKERACDIALLWMDSVNLS---IRSYSLNHIMGRIEDVESGKYWSITGIYGYPDET 120
GK++ IA+L+ D++ IR H + + + + SI IY P+
Sbjct: 63 -QANGKQKKAGIAILFADAIGFKPTKIRKDKDGHFIFVKGNTQYDEI-SIINIYA-PNHN 119
Query: 121 NKKKTWDLLRSLTPVEEDMWLCCGDFNDILSNEEKAGGNIRSFLQLSLSRNAVEDCNLLD 180
+ + L ++ + + GDFN L+ +++ S L L+ + ++ +L D
Sbjct: 120 APQFIRETLTDMSNLISSTSIVVGDFNTPLAVLDRSSKKKLSKEILDLN-STIQHLDLTD 178
Query: 181 LGFNGYP----FTWSNGRHGSGRI*CRLDRVFASDEFIKRFSPIQINHLARFGSDHAAIS 236
+ +P +T+ + HG+ ++D + + +F I+I + SDH I
Sbjct: 179 IYRTFHPNKTEYTFFSSAHGTYS---KIDHILGHKSNLSKFKKIEI--IPCIFSDHHGIK 233
Query: 237 IELDVCLE-DAHKK--KLHVFRFEKCWADDDRCEAMVR-------------GSWNNATG- 279
+EL+ H K KL+ + W D+ + + + W+ A
Sbjct: 234 VELNNNRNLHTHTKTWKLNNLMLKDTWVIDEIKKEITKFLEQNNNQDTNYQNLWDTAKAV 293
Query: 280 LASKIEAMQSLDSKFKEYITSEIRKELLEIETQLNNHASWDGSPDNITKFRELEAKHSEL 339
L K A+Q+ K + + + L ++E + +++ P + ++ A+ +E+
Sbjct: 294 LRGKFIALQAFLKKTEREEVNNLMGHLKQLEKEEHSNPK----PSRRKEITKIRAELNEI 349
Query: 340 LQTEETMWRQRSRATWLKEGDKNSKFFHAKAKQRGKINSIKKIKDDRGVWWNGRDKVEKV 399
+S++ + ++ +K K +++ + I I++ +++K+
Sbjct: 350 ENKRIIQQINKSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKI 409
Query: 400 LVDYFAGLFTSS--NPTTVDQTCLVVK-DRLSPEHVEWCNRSFSPAEIKDAIDQMHPLKA 456
L +Y+ L++ N +DQ RLS + VE NR S +EI I + K+
Sbjct: 410 LNEYYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNLPKKKS 469
Query: 457 PGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIALIPKC-KNPSSPNQF 515
PGPDG + F+Q + + +++L I EG + I LIPK K+P+ +
Sbjct: 470 PGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENY 529
Query: 516 RPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGRLITDNAIIAMECFHWMKKKT 575
RPISL N+ KI+ K + NR++ + +I+ +Q F+ G N ++ + K
Sbjct: 530 RPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLK 589
Query: 576 KGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCISTVSYQILINGQPS 635
K M + +D KA+D I+ F+ L+ + + I S + I++NG
Sbjct: 590 N--KDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKL 647
Query: 636 RRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIARNAPKITHLLFADD 695
+ F G RQG PLSP LF + VL+ +++E+ I GI I K++ LFADD
Sbjct: 648 KSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREEK---AIKGIHIGSEEIKLS--LFADD 702
Query: 696 GLLFARASVEEAQCIISVLNTYQDASGQIVNLDKSEVSYSRNVLNHDKEIICQRINIKTV 755
+++ + + ++ V+ Y + SG +N KS V++ N ++ + I V
Sbjct: 703 MIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKS-VAFIYTNNNQAEKTVKDSIPFTVV 761
Query: 756 NTHSRYLGLPVIFGRSK--KEVFSFVQERVWKKIKGWKEKCLSRAGKETLIK--AVAQAI 811
+YLG+ + KE + +++ + + + WK S G+ ++K + +AI
Sbjct: 762 PKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAI 821
Query: 812 PNYIMSCYKLPEGCCDAVEGMLAKFWWASDEYKRKIHWMRWGRLGETKKRGGLGFRSFSD 871
N+ K P +E ++ F W +K + L K GG+
Sbjct: 822 YNFNAIPIKAPLSYFKDLEKIILHFIW-----NQKKPQIAKTLLSNKNKAGGITLPDLRL 876
Query: 872 FNKALLGKQYW 882
+ K+++ K W
Sbjct: 877 YYKSIVIKTAW 887
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 176 bits (447), Expect = 3e-43
Identities = 206/918 (22%), Positives = 393/918 (42%), Gaps = 80/918 (8%)
Query: 6 LLSWNCRGLGNPRAIRALLRLIRIQNPQVVFLMETRLKSDEMERVRINCGFSSGLFVPCD 65
+L+ N GL P L I+ Q+P V + ET L + R++I G
Sbjct: 9 ILTLNVNGLNAPIKRHRLANWIKSQDPSVCCIQETHLTCRDTHRLKIK-----GWRNIYQ 63
Query: 66 GFGKERACDIALLWMDSVNLS---IRSYSLNH---IMGRIEDVESGKYWSITGIYGYPDE 119
GK++ +A+L D + I+ H + G I+ E +I IY P+
Sbjct: 64 ANGKQKKAGVAILVSDKTDFKPTKIKRDKEGHYIMVKGSIQQEEL----TILNIYA-PNT 118
Query: 120 TNKKKTWDLLRSLTPVEEDMWLCCGDFNDILSNEEKAGGNIRSFLQLSLSRNAVEDCNLL 179
+ +L L + + GDFN LS +++ + + +A+ +L+
Sbjct: 119 GAPRFIKQVLSDLQRDLDSHTIIMGDFNTPLSTLDRSTRQ-KINKDIQELNSALHQADLI 177
Query: 180 DLGFNGYP----FTWSNGRHGSGRI*CRLDRVFASDEFIKRFSPIQINHLARFGSDHAAI 235
D+ +P +T+ + H + + D + S + + +I + SDH+AI
Sbjct: 178 DIYRTLHPKSTEYTFFSAPHHTYS---KTDHILGSKTLLSKCKRTEI--ITNCLSDHSAI 232
Query: 236 SIEL----------------DVCLEDAHKKKLHVFRFEKCWADDDRCEAMVRGSWNNATG 279
+EL ++ L D +K + ++ + + W+ A
Sbjct: 233 KLELRIKKLTQNHSTTWKLNNLLLNDYWVHNEMKAEIKKFFETNENKDTTYQNLWDTAKA 292
Query: 280 LA-SKIEAMQSLDSKFKEYITSEIRKELLEIETQLNNHASWDGSPDNITKFRELEAKHSE 338
+ K A+ + K + + +L E+E Q ++ + I EL+
Sbjct: 293 VCRGKFIALNAHKRKQERSKIDTLISQLKELEKQEQTNSKASRRQEIIKIRAELKE---- 348
Query: 339 LLQTEETMWRQRSRATWLKEG-DKNSKFFHAKAKQRGKINSIKKIKDDRGVWWNGRDKVE 397
++T++T+ + +W E +K + K++ + N I IK+DRG +++
Sbjct: 349 -IETQKTLQKINESRSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQ 407
Query: 398 KVLVDYFAGLFTSS--NPTTVDQTC-LVVKDRLSPEHVEWCNRSFSPAEIKDAIDQMHPL 454
+ +Y+ L+ + N +D+ RL+ E VE NR + +EI+ I+ +
Sbjct: 408 TTIREYYKHLYANKLENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNK 467
Query: 455 KAPGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIALIPK-CKNPSSPN 513
K+PGP+G A F+Q+Y + ++ L I EG + I LIPK ++ +
Sbjct: 468 KSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKE 527
Query: 514 QFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFV---QGRLITDNAIIAMECFHW 570
FRPISL N+ KI+ K +AN+++ + +++ +Q F+ QG +I ++ +
Sbjct: 528 NFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINR 587
Query: 571 MKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCISTVSYQILI 630
K M + +D KA+D+I+ F+ L + + I + I++
Sbjct: 588 TKDTNH-----MIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIIL 642
Query: 631 NGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIARNAPKITHL 690
NGQ + G RQG PLSP L + VL+ +++E+ +I GI++ + K++
Sbjct: 643 NGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIRQEK---EIKGIQLGKEEVKLS-- 697
Query: 691 LFADDGLLFARASVEEAQCIISVLNTYQDASGQIVNLDKSEVSYSRNVLNHDKEIICQRI 750
LFADD +++ + AQ ++ +++ + SG +N+ KS+ N + +I+ + +
Sbjct: 698 LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSE-L 756
Query: 751 NIKTVNTHSRYLGLPVIFGRSKKEVFSFVQERVWKKIK----GWKEKCLSRAGKETLIK- 805
+ +YLG+ + R K++F + + +IK WK S G+ ++K
Sbjct: 757 PFTIASKRIKYLGIQLT--RDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKM 814
Query: 806 -AVAQAIPNYIMSCYKLPEGCCDAVEGMLAKFWWASDEYKRKIHWMRWGRLGETKKRGGL 864
+ + I + KLP +E KF W +++ H + L + K GG+
Sbjct: 815 AILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIW----NQKRAHIAK-STLSQKNKAGGI 869
Query: 865 GFRSFSDFNKALLGKQYW 882
F + KA + K W
Sbjct: 870 TLPDFKLYYKATVTKTAW 887
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 172 bits (436), Expect = 6e-42
Identities = 213/933 (22%), Positives = 399/933 (41%), Gaps = 110/933 (11%)
Query: 6 LLSWNCRGLGNPRAIRALLRLIRIQNPQVVFLMETRLKSDEMERVRINCGFSSGLFVPCD 65
L+S N GL +P L + Q+P L ET L+ + +R+ G
Sbjct: 36 LISLNINGLNSPIKRHRLTDWLHKQDPTFCCLQETHLREKDRHYLRVK-----GWKTIFQ 90
Query: 66 GFGKERACDIALLWMDSVNLS---IRSYSLNH---IMGRIEDVESGKYWSITGIYGYPDE 119
G ++ +A+L D ++ I+ H I G+I E SI IY P+
Sbjct: 91 ANGLKKQAGVAILISDKIDFQPKVIKKDKEGHFILIKGKILQEEL----SILNIYA-PNA 145
Query: 120 TNKKKTWDLLRSLTPVEEDMWLCCGDFNDILSNEEKAGGNIRSFLQLSLSRNAVEDCNLL 179
D L L + GDFN LS+++++ + + L+ ++ +L
Sbjct: 146 RAATFIRDTLVKLKAYIAPHTIIVGDFNTPLSSKDRSWKQKLNRDTVKLTE-VMKQMDLT 204
Query: 180 DLGFNGYP----FTWSNGRHGSGRI*CRLDRVFASDEFIKRFSPIQINHLARFGSDHAAI 235
D+ YP +T+ + HG+ ++D + + R+ I+I + SDH +
Sbjct: 205 DIYRTFYPKTKGYTFFSAPHGTFS---KIDHIIGHKTGLNRYKNIEI--VPCILSDHHGL 259
Query: 236 SIEL---------------------DVCLEDAHKKKLHVF---------RFEKCWADDDR 265
+ D +++ KK++ F + W D
Sbjct: 260 RLIFNNNINNGKPTFTWKLNNTLLNDTLVKEGIKKEIKDFLEFNENEATTYPNLW---DT 316
Query: 266 CEAMVRGSWNNATGLASKIEAMQSLDSKFKEYITSEIRKELLEIETQLNNHASWDGSPDN 325
+A +RG K+ A+ + K + TS + L +E + N SP
Sbjct: 317 MKAFLRG----------KLIALSASKKKRETAHTSSLTTHLKALEKKEAN------SPKR 360
Query: 326 ITKFRELEAKHSEL--LQTEETMWRQRSRATWLKEGDKNSKFFHAKAK----QRGKINSI 379
++ +E+ E+ ++T T+ R +W E K +K A+ R KI I
Sbjct: 361 -SRRQEIIKLRGEINQVETRRTIQRINQTRSWFFE--KINKIDKPLARLTKGHRDKI-LI 416
Query: 380 KKIKDDRGVWWNGRDKVEKVLVDYFAGLFTSSNPTTVDQTCLVVK---DRLSPEHVEWCN 436
KI++++G ++++ + ++ L+++ + + + +L+ + V+ N
Sbjct: 417 NKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLN 476
Query: 437 RSFSPAEIKDAIDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECIN 496
SP EI+ I+ + K+PGPDG A F+Q + + + L I EG
Sbjct: 477 SPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFY 536
Query: 497 KTFIALIPKC-KNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGR 555
+ I LIPK K+P+ FRPISL N+ KI+ K +ANR++ + I+ +Q F+ G
Sbjct: 537 EATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGM 596
Query: 556 LITDNAIIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIIN 615
N ++ H++ K K M + LD KA+D+I+ F+ +L+ +N
Sbjct: 597 QGWFNIRKSINVIHYINKLKD--KNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLN 654
Query: 616 TILQCISTVSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKI 675
I S I +NG+ + G RQG PLSPYLF + VL+ +++++ +I
Sbjct: 655 MIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQK---EI 711
Query: 676 HGIRIARNAPKITHLLFADDGLLFARASVEEAQCIISVLNTYQDASGQIVNLDKSEVSYS 735
GI+I + KI+ L ADD +++ + +++++N++ + G +N +KS
Sbjct: 712 KGIQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLY 769
Query: 736 RNVLNHDKEIICQRINIKTVNTHSRYLGLPVIFGRSKKEV----FSFVQERVWKKIKGWK 791
+KE I + V + +YLG V + K++ F +++ + + ++ WK
Sbjct: 770 TKNKQAEKE-IRETTPFSIVTNNIKYLG--VTLTKEVKDLYDKNFKSLKKEIKEDLRRWK 826
Query: 792 EKCLSRAGKETLIK--AVAQAIPNYIMSCYKLPEGCCDAVEGMLAKFWWASDEYKRKIHW 849
+ S G+ ++K + +AI + K+P + +EG + KF W + + +
Sbjct: 827 DLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKKPR----- 881
Query: 850 MRWGRLGETKKRGGLGFRSFSDFNKALLGKQYW 882
+ L + + GG+ + +A++ K W
Sbjct: 882 IAKSLLKDKRTSGGITMPDLKLYYRAIVIKTAW 914
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 159 bits (401), Expect = 6e-38
Identities = 210/897 (23%), Positives = 365/897 (40%), Gaps = 73/897 (8%)
Query: 4 MKLLSWNCRGLGNPRAIRALLRLIRIQNPQVVFLMETRLKSDEMERVRINCGFSSGLFVP 63
+ + + N G NP + +L +R V FL ET + E+E N + +F
Sbjct: 3 LSISTLNTNGCRNPFRMFQVLSFLRQGGYSVSFLQETHT-TPELE-ASWNLEWKGRVFF- 59
Query: 64 CDGFGKERACDIALLWMDSVNLSIRSYSLNHIMGRIEDV---ESGKYWSITGIYGYPDET 120
+C + L+ DS + S + + I GR+ + ESG+ +++ +Y
Sbjct: 60 --NHLTWTSCGVVTLFSDSFQPEVLS-ATSVIPGRLLHLRVRESGRTYNLMNVYAPTTGP 116
Query: 121 NKKKTWDLLRSL--TPVEEDMWLCCGDFNDILSNEEKAGGNIRSFLQLSLSRNAVEDCNL 178
+ + ++ L + T ++ + GDFN L ++ R + S+ R + +L
Sbjct: 117 ERARFFESLSAYMETIDSDEALIIGGDFNYTLDARDRNVPKKRDSSE-SVLRELIAHFSL 175
Query: 179 LDLGFNGYP----FTWSNGRHGSGRI*CRLDRVFASDEFIKRFSPIQINHLARFGSDHAA 234
+D+ P FT+ R G R+DR++ S + R I LA F SDH
Sbjct: 176 VDVWREQNPETVAFTYVRVRDGHVSQ-SRIDRIYISSHLMSRAQSSTIR-LAPF-SDHNC 232
Query: 235 ISIELDVCLEDAHKKKLHVFRFEKCWADDDRCEAMVRGSWNNATGLASKIEAMQS----- 289
+S+ + + K + F +D+ VR +W + +
Sbjct: 233 VSLRMSIA---PSLPKAAYWHFNNSLLEDEGFAKSVRDTWRGWRAFQDEFATLNQWWDVG 289
Query: 290 ---LDSKFKEYITSEIRKELLEIETQ----LNNHASWDGSPDNITKFRELEAKHSELLQT 342
L +EY S + EIE L+ GS D + LE K + L
Sbjct: 290 KVHLKLLCQEYTKSVSGQRNAEIEALNGEVLDLEQRLSGSEDQALQCEYLERKEA-LRNM 348
Query: 343 EETMWRQ---RSRATWLKEGDKNSKFFHAKAKQRGKINSIKKIKDDRGVWWNGRDKVEKV 399
E+ R RSR L + D+ S+FF+A K++G I + + G + +
Sbjct: 349 EQRQARGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDR 408
Query: 400 LVDYFAGLFTSSNPTTVDQTCLVVKDRL---SPEHVEWCNRSFSPAEIKDAIDQMHPLKA 456
++ LF S +P + D C + D L S E + E+ A+ M K+
Sbjct: 409 ARSFYQNLF-SPDPISPD-ACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKS 466
Query: 457 PGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIALIPKCKNPSSPNQFR 516
PG DGL FFQ +W +G + + +G+ + ++L+PK + +R
Sbjct: 467 PGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWR 526
Query: 517 PISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGRLITDNAIIAMECFHWMKKKTK 576
P+SL + KIV K I+ RLK +L E++ +QS V GR I DN + + H+ ++
Sbjct: 527 PVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRT-- 584
Query: 577 GKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCISTVSYQILINGQPSR 636
+ + LD KA+DR++ ++ LQ+ +F + + ++ + IN +
Sbjct: 585 -GLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTA 643
Query: 637 RFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIARNAPKITHLLFADDG 696
RG+RQG PLS L+ L LL+K ++ G+ + ++ +ADD
Sbjct: 644 PLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRK-----RLTGLVLKEPDMRVVLSAYADDV 698
Query: 697 LLFAR--ASVEEAQCIISVLNTYQDASGQIVNLDKSEVSYSRNVLNHDKEIICQRINIKT 754
+L A+ +E AQ Y AS +N KS +L ++ +
Sbjct: 699 ILVAQDLVDLERAQ---ECQEVYAAASSARINWSKSS-----GLLEGSLKVDFLPPAFRD 750
Query: 755 VNTHS---RYLGLPVIFGR-SKKEVFSFVQERVWKKIKGWK--EKCLSRAGKETLIKAVA 808
++ S +YLG+ + + F ++E V ++ WK K LS G+ +I +
Sbjct: 751 ISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLV 810
Query: 809 QAIPNYIMSCYKLPEGCCDAVEGMLAKFWWASDEYKRKIHWMRWGRLGETKKRGGLG 865
+ Y + C + ++ L F W HW+ G K GG G
Sbjct: 811 ASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGK------HWVSAGVSSLPLKEGGQG 861
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 129 bits (324), Expect = 5e-29
Identities = 61/143 (42%), Positives = 93/143 (64%), Gaps = 1/143 (0%)
Query: 810 AIPNYIMSCYKLPEGCCDAVEGMLAKFWWASDEYKRKIHWMRWGRLGETKKR-GGLGFRS 868
A+P Y MSC++L + C + + +FWW+S E KRKI W+ W +L ++K+ GGLGFR
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 869 FSDFNKALLGKQYWRLLADNNSLMSSILKSRYFPRTSISITKVGFQPRYAWRSLMKAREL 928
FN+ALL KQ +R++ ++L+S +L+SRYFP +S+ VG +P YAWRS++ REL
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 929 VENDAHWVIGNGRTVRIFKDNWI 951
+ IG+G +++ D WI
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRWI 144
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 90.5 bits (223), Expect = 3e-17
Identities = 40/67 (59%), Positives = 51/67 (75%)
Query: 629 LINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIARNAPKIT 688
+ING P P RGLRQGDPLSPYLFILC VLSGL ++ ++ ++ GIR++ N+P+I
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 689 HLLFADD 695
HLLFADD
Sbjct: 73 HLLFADD 79
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 79.7 bits (195), Expect = 5e-14
Identities = 86/335 (25%), Positives = 139/335 (40%), Gaps = 27/335 (8%)
Query: 374 GKINSIKKIKDDRGVWWNGRDKVEKVLVDYF-----AGLF-TSSNPTTVDQTCLVVKDRL 427
G+ + IK+ W K +++D AGL T ++ + C +
Sbjct: 246 GETQRRQTIKESNNSWKKNMSKAAHIVLDGDTDACPAGLEGTEASGAIMRAGCPTTRHLR 305
Query: 428 SPEHVEWCN--RSFSPAEIKDAIDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGI 485
S E N R S EIK+ + A GPDG+ W + + SL I
Sbjct: 306 SRMQGEIKNLWRPISNDEIKEV--EACKRTAAGPDGMTTTA----WNSIDECIKSLFNMI 359
Query: 486 LNEGKSTECINKTFIALIPKCKNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMD 545
+ G+ + LIPK P FRP+S+ +V ++ + +ANR+ ++D
Sbjct: 360 MYHGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLD 417
Query: 546 EEQSAFVQGRLITDNAIIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQ 605
Q AF+ + +N + + K KG + LD+ KA+D +E + L+
Sbjct: 418 TRQRAFIVADGVAENTSLLSAMIKEARMKIKG---LYIAILDVKKAFDSVEHRSILDALR 474
Query: 606 SMNFPENIINTILQCISTVSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGL 665
P + N I+ ++ + R P RG+RQGDPLSP LF V+ +
Sbjct: 475 RKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNC---VMDAV 531
Query: 666 LKKEEKDNKIHGIRIARNAPKITHLLFADDGLLFA 700
L++ ++ A KI L+FADD +L A
Sbjct: 532 LRRLPENT-----GFLMGAEKIGALVFADDLVLLA 561
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 74.3 bits (181), Expect = 2e-12
Identities = 82/344 (23%), Positives = 142/344 (40%), Gaps = 30/344 (8%)
Query: 391 NGRDKV----EKVLVDYFAGLFTSSNPTTVDQTCLVVKDRLSPEHVEWCNRSFSPAEIKD 446
NG D+ ++++V Y+ + T +P++ + + L R +S +D
Sbjct: 297 NGTDESVMPSQEIMVPYWREVMTQPSPSSCSGEVIQMDHSLE--------RVWSAITEQD 348
Query: 447 A-IDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIALIPK 505
++ +PGPDG+ ++ + +++L L N S FI PK
Sbjct: 349 LRASRVSLSSSPGPDGITPKSAREVPSGIMLRIMNLILWCGNLPHSIRLARTVFI---PK 405
Query: 506 CKNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGRLITDNAIIAM 565
P FRPIS+ +V+++ + +A RL + D Q F+ DNA I
Sbjct: 406 TVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGFLPTDGCADNATIVD 463
Query: 566 ECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCISTVS 625
++ K + LD++KA+D + + + L++ P+ ++ +
Sbjct: 464 LV---LRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGG 520
Query: 626 YQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIARNAP 685
+ +G S F P RG++QGDPLSP LF L + L L E + I A
Sbjct: 521 TSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIGAKVGNAITNA---- 576
Query: 686 KITHLLFADDGLLFARASVEEAQCIISVLNTYQDASGQIVNLDK 729
FADD +LFA + Q ++ + G +N DK
Sbjct: 577 ----AAFADDLVLFAETRM-GLQVLLDKTLDFLSIVGLKLNADK 615
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 73.9 bits (180), Expect = 3e-12
Identities = 63/230 (27%), Positives = 108/230 (46%), Gaps = 13/230 (5%)
Query: 502 LIPKCKNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGRLITDNA 561
LIPK + +P+ +RPI++ + + +++ + +A RL+ + ++ A + G L+
Sbjct: 63 LIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQKGYARIDGTLVNSLL 122
Query: 562 IIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCI 621
+ ++ + + +K + LD+ KA+D + S + LQ + E N I +
Sbjct: 123 LDT-----YISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITGSL 177
Query: 622 STVSYQILIN-GQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRI 680
S + I + G +R+ RG++QGDPLSP+LF VL LL + I G
Sbjct: 178 SDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLF---NAVLDELLCSLQSTPGIGG--- 231
Query: 681 ARNAPKITHLLFADDGLLFARASVEEAQCIISVLNTYQDASGQIVNLDKS 730
KI L FADD LL V + +V N ++ G +N KS
Sbjct: 232 TIGEEKIPVLAFADDLLLLEDNDVLLPTTLATVANFFR-LRGMSLNAKKS 280
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 64.7 bits (156), Expect = 2e-09
Identities = 42/161 (26%), Positives = 76/161 (47%), Gaps = 4/161 (2%)
Query: 503 IPKCKNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAFVQGRLITDNAI 562
IPK NPSSP+ +RPISL + +I+ + I +R++ ++ Q F+ R + +
Sbjct: 673 IPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLV 732
Query: 563 IAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCIS 622
++ +H + K +K + + D AKA+D++ + L + + + +
Sbjct: 733 RSISLYHSI---LKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLH 789
Query: 623 TVSYQILINGQPSRRFFP-ERGLRQGDPLSPYLFILCANVL 662
++ + IN S +P G+ QG P LFIL N L
Sbjct: 790 LRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDL 830
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 59.3 bits (142), Expect = 7e-08
Identities = 67/273 (24%), Positives = 116/273 (41%), Gaps = 25/273 (9%)
Query: 431 HVEWCNRSFSPAEIKDAIDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGI-LNEG 489
H+E S EIK A + K GPDG+ + W + L I + G
Sbjct: 151 HMETLWDPVSLIEIKSA--RASNEKGAGPDGVTP----RSWNALDDRYKRLLYNIFVFYG 204
Query: 490 KSTECINKTFIALIPKCKNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQS 549
+ I + PK + P FRP+S+C+V+++ K +A R + DE Q+
Sbjct: 205 RVPSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRF--VSCYTYDERQT 262
Query: 550 AF--VQGRLITDNAIIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSM 607
A+ + G I + + A+ + + + +K + LD+ KA++ + S + +
Sbjct: 263 AYLPIDGVCINVSMLTAI-----IAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEA 317
Query: 608 NFPENIINTILQCISTVSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLK 667
P +++ I + V ++ G+ G+ QGDPLS LF L L
Sbjct: 318 GCPPGVVDYIADMYNNVITEMQFEGKCELASI-LAGVYQGDPLSGPLFTLAYEKALRALN 376
Query: 668 KEEKDNKIHGIRIARNAPKITHLLFADDGLLFA 700
E + + I +R+ +A ++DDGLL A
Sbjct: 377 NEGRFD-IADVRVNASA-------YSDDGLLLA 401
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 57.8 bits (138), Expect = 2e-07
Identities = 42/168 (25%), Positives = 77/168 (45%), Gaps = 9/168 (5%)
Query: 562 IIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCI 621
II +E H++K + K + LD+ KA+D + + +++ + + + I+ I
Sbjct: 6 IIMLE--HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTI 63
Query: 622 STVSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLKKEEKDNKIHGIRIA 681
+ I++ G+ + + + G++QGDPLSP LF + + L L E+ +
Sbjct: 64 TDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM------ 117
Query: 682 RNAPKITHLLFADDGLLFARASVEEAQCIISVLNTYQDASGQIVNLDK 729
A KI L FADD LL ++ + + Y G +N +K
Sbjct: 118 TPACKIASLAFADDLLLLEDRDIDVPNSLATTC-AYFRTRGMTLNPEK 164
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 55.8 bits (133), Expect = 8e-07
Identities = 74/347 (21%), Positives = 132/347 (37%), Gaps = 60/347 (17%)
Query: 443 EIKDAIDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIAL 502
E+ + ++ ++PG DG+ + W + + SL + CI +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLF---------SRCIRLGYFPA 488
Query: 503 IPKC-----------KNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEIMDEEQSAF 551
KC K+ P+ +R I L V K++ + NR++ +LPE Q F
Sbjct: 489 EWKCPRVVSLLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPEGC-RWQFGF 547
Query: 552 VQGRLITDNAIIAMECFHWMKKKT----KGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSM 607
QGR + D W K+ + V+ +D A+D +EWS S L +
Sbjct: 548 RQGRCVED---------AWRHVKSSVGASAAQYVLGTFVDFKGAFDNVEWSAALSRLADL 598
Query: 608 NFPENIINTILQCISTVSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLLK 667
E + Q + ++ + + RG QG P+++ + +VL
Sbjct: 599 GCRE---MGLWQSFFSGRRAVIRSSSGTVEVPVTRGCPQGSISGPFIWDILMDVL----- 650
Query: 668 KEEKDNKIHGIRIARNAPKITHLLFADDGLLF----ARASVEEAQC-IISVLNTYQDASG 722
+ R P +ADD LL +RA +EE ++S++ T+ G
Sbjct: 651 ------------LQRLQPYCQLSAYADDLLLLVEGNSRAVLEEKGAQLMSIVETWGAEVG 698
Query: 723 QIVNLDKSEVSYSRNVLNHDKEIICQRINIKTVNTHSRYLGLPVIFG 769
++ K+ + + L + N+ V + RYLG+ V G
Sbjct: 699 DCLSTSKTVIMLLKGALRRAPTVRFAGRNLPYVRS-CRYLGITVSEG 744
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 43.1 bits (100), Expect = 0.005
Identities = 65/324 (20%), Positives = 127/324 (39%), Gaps = 19/324 (5%)
Query: 358 EGDKNSKFFHAKAKQRGKINSIKKIKDDRGVWWNGRDKVEKVLVDYFAGLFTSSNPTTVD 417
+G N + + + + I++ G W + +V ++ F +
Sbjct: 357 DGSTNFSLWRITKRYKTQATPNSAIRNPAGGWCRTSREKTEVFANHLEQRFKALAFAPES 416
Query: 418 QTCLVVKDRLSPEHVEWCNRSFSPAEIKDAIDQMHPLKAPGPDGLP-----ALFFQKYWY 472
+ +V + +P + + E+K+ + ++ P KAPG D L L Q Y
Sbjct: 417 HSLMVAESLQTPFQMALPADPVTLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLY 476
Query: 473 IVGSEVVSLALGILNEGKSTECINKTFIALIPKCKNPSSPNQFRPISLCNVVMKIVTKTI 532
+V L +G + + T I I ++ K P + +RP SL + K++ + I
Sbjct: 477 LVLIFNSILRVGYFPKARPTASI----IMILKPGKQPLDVDSYRPTSLLPSLGKMLERLI 532
Query: 533 ANRLKPILPEIMDEEQSAFVQG-RLITDNAIIAMECFHWMKKKTKGKKGVMAMKLDMAKA 591
NR+ + E + F G RL ++ + + K+ + LD+ +A
Sbjct: 533 LNRI--LTSEEVTRAIPKFQFGFRLQHGTPEQLHRVVNFALEALEKKEYAGSCFLDIQQA 590
Query: 592 YDRIEWSFVKSMLQSMNFPENIINTILQCISTVSYQILINGQPSRRFFPERGLRQGDPLS 651
+DR+ + +S+ P+ + I + + +G S F E G+ QG L
Sbjct: 591 FDRVWHPGLLYKAKSLLSPQ-LFQLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLG 649
Query: 652 PYLFILCA------NVLSGLLKKE 669
P L+ + N ++GL + E
Sbjct: 650 PTLYSIFTADMPNQNAVTGLAEGE 673
>APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (Class II
apurinic/apyrimidinic(AP)-endonuclease)
Length = 361
Score = 43.1 bits (100), Expect = 0.005
Identities = 55/261 (21%), Positives = 102/261 (39%), Gaps = 28/261 (10%)
Query: 4 MKLLSWNCRGLGNPRAIRALLRLIRIQNPQVVFLMETRLKSDEMERVRINCGFSSGLFVP 63
MK++SWN G + + + + +NP V+ L ET++ +++ ++ G+ F+
Sbjct: 105 MKIISWNVAGFKSVLS-KGFTECVEKENPDVLCLQETKINPSNIKKDQMPKGYEYH-FIE 162
Query: 64 CDGFGKERACDIALLWMDSVNLSIRSYSLNHIMGRIEDVESGKYWSI------TGIYGYP 117
D G + +++ I + + GR+ +E +++ + G G
Sbjct: 163 ADQKGHHGTGVLTKKKPNAITFGI-GIAKHDNEGRVITLEYVQFYIVNTYIPNAGTRGLQ 221
Query: 118 DETNKKKTWDL-----LRSLTPVEEDMWLCCGDFN---------DILSNEEKAGGNIRSF 163
+ K WD+ L L + +W CGD N + +N++ AG I
Sbjct: 222 RLDYRIKEWDVDFQAYLEKLNATKPIIW--CGDLNVAHTEIDLKNPKTNKKSAGFTIEER 279
Query: 164 LQLS--LSRNAVEDCNLLDLGFNGYPFTWSN-GRHGSGRI*CRLDRVFASDEFIKRFSPI 220
S L + V+ + G G WS G S + RLD S +
Sbjct: 280 TSFSNFLEKGYVDSYRHFNPGKEGSYTFWSYLGGGRSKNVGWRLDYFVVSKRLMDSIKIS 339
Query: 221 QINHLARFGSDHAAISIELDV 241
+ + GSDH I + +D+
Sbjct: 340 PFHRTSVMGSDHCPIGVVVDL 360
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 42.0 bits (97), Expect = 0.011
Identities = 36/158 (22%), Positives = 66/158 (40%), Gaps = 8/158 (5%)
Query: 443 EIKDAIDQMHPLKAPGPDGLPALFFQKYWYIVGSEVVSLALGILNEGKSTECINKTFIAL 502
E+ D++ + K+PGPDG+ + W + + L L E + + +
Sbjct: 409 EVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLVI 468
Query: 503 IPKC--KNPSSPNQFRPISLCNVVMKIVTKTIANRLKPILPEI-MDEEQSAFVQGRLITD 559
+ K + S P +RPI L + + K++ + RL L ++ + Q AF G+ D
Sbjct: 469 LLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTED 528
Query: 560 NAIIAMECFHWMKKKTKGKKGVMAMKLDMAKAYDRIEW 597
A C + K V+ + +D A+D + W
Sbjct: 529 ----AWRCVQ-RHVECSEMKYVIGLNIDFQGAFDNLGW 561
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 41.2 bits (95), Expect = 0.019
Identities = 87/435 (20%), Positives = 170/435 (39%), Gaps = 32/435 (7%)
Query: 246 AHKKKLHVFRFEKCWADDDRCEAMVRGSWNNATGL-ASKIEAMQSLDSKFKEYITSEIRK 304
AH +++F+ + + + +NAT L SK+ L + + R
Sbjct: 236 AHNADINIFKTHLEQLSEVNMQILEAVDIDNATSLFMSKLSEAAQLAAPRNRHEVEAFRP 295
Query: 305 --------ELLEIETQLNNHASWDGSPDNITKFRELEAKHSELLQTEETMWRQRSRATWL 356
LL ++ ++ + G P + +++ ++ + L +Q T+L
Sbjct: 296 LQLPSSILALLRLKRRVRKEYARTGDP----RMQQIHSRLANCLHKALARRKQAQIDTFL 351
Query: 357 KE--GDKNSKFFHAKAKQRGKINSIKK--IKDDRGVWWNGRDKVEKVLVDYFAGLFTSSN 412
D ++ + + +R K K IK+ G W + +V + FT N
Sbjct: 352 DNLGADASTNYSLWRITKRFKAQPTPKSAIKNPSGGWCRTSLEKTEVFANNLEQRFTPYN 411
Query: 413 PTTVDQTCLVVKDRL-SPEHVEWCNRSFSPAEIKDAIDQMHPLKAPGPDGLPALFFQKYW 471
+ C V++ L SP + + + E+K+ I ++ KAPG D L +
Sbjct: 412 YAP-ESLCRQVEEYLESPFQMSLPLSAVTLEEVKNLIAKLPLKKAPGEDLLDNRTIR--- 467
Query: 472 YIVGSEVVSLAL---GILNEGKSTECINKTFIALIPKC-KNPSSPNQFRPISLCNVVMKI 527
+ + LAL +L+ G + I +I K K P+ + +RP SL + KI
Sbjct: 468 LLPDQALQFLALIFNSVLDVGYFPKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKI 527
Query: 528 VTKTIANRLKPI--LPEIMDEEQSAFVQGRLITDNAIIAMECFHWMKKKTKGKKGVMAMK 585
+ + I NRL + + + + Q F RL ++ + + K+ +
Sbjct: 528 MERLILNRLLTCKDVTKAIPKFQFGF---RLQHGTPEQLHRVVNFALEAMENKEYAVGAF 584
Query: 586 LDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCISTVSYQILINGQPSRRFFPERGLR 645
LD+ +A+DR+ W FP + + + ++ + ++G S G+
Sbjct: 585 LDIQQAFDRV-WHPGLLYKAKRLFPPQLYLVVKSFLEERTFHVSVDGYKSSIKPIAAGVP 643
Query: 646 QGDPLSPYLFILCAN 660
QG L P L+ + A+
Sbjct: 644 QGSVLGPTLYSVFAS 658
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 36.2 bits (82), Expect = 0.63
Identities = 25/103 (24%), Positives = 50/103 (48%), Gaps = 2/103 (1%)
Query: 565 MECFH-WMKKKTKGKKGVMAMKLDMAKAYDRIEWSFVKSMLQSMNFPENIINTILQCIST 623
+EC + W K +G + + LD +KA+D++ + L S+ +++I + ++
Sbjct: 2 LECMNNWTKNIDEGAQTDVKY-LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTN 60
Query: 624 VSYQILINGQPSRRFFPERGLRQGDPLSPYLFILCANVLSGLL 666
S+++ + S G+ QG +SP LF + N +S L
Sbjct: 61 RSFKVKVGNTLSEPKKTVCGVPQGSVISPVLFGIFVNEISANL 103
>UBPG_YEAST (Q02863) Ubiquitin carboxyl-terminal hydrolase 16 (EC
3.1.2.15) (Ubiquitin thiolesterase 16)
(Ubiquitin-specific processing protease 16)
(Deubiquitinating enzyme 16)
Length = 499
Score = 35.0 bits (79), Expect = 1.4
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query: 293 KFKEYITSEIRKELLEIETQLNNHASWDG-SPDNITKFRELEAKHSELLQTEETM 346
+F EY+ IR LLE+ET+L+N+A D + DN T+ LE + L E++
Sbjct: 76 RFVEYL-KRIRTVLLELETKLSNNAKGDNPTVDNTTRHSRLENSSNSLAPLHESL 129
>M6P1_HUMAN (O60664) Mannose-6-phosphate receptor binding protein 1
(Cargo selection protein TIP47) (47 kDa mannose
6-phosphate receptor-binding protein) (47 kDa
MPR-binding protein) (Placental protein 17)
Length = 434
Score = 34.3 bits (77), Expect = 2.4
Identities = 16/49 (32%), Positives = 27/49 (54%)
Query: 700 ARASVEEAQCIISVLNTYQDASGQIVNLDKSEVSYSRNVLNHDKEIICQ 748
AR VE+ Q S ++++QD S I+ + V+ +R L+H E + Q
Sbjct: 362 ARRQVEDLQATFSSIHSFQDLSSSILAQSRERVASAREALDHMVEYVAQ 410
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.137 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,946,046
Number of Sequences: 164201
Number of extensions: 7234706
Number of successful extensions: 15067
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 15016
Number of HSP's gapped (non-prelim): 40
length of query: 1363
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1240
effective length of database: 39,777,331
effective search space: 49323890440
effective search space used: 49323890440
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146790.3


