

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.3 - phase: 0
(348 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
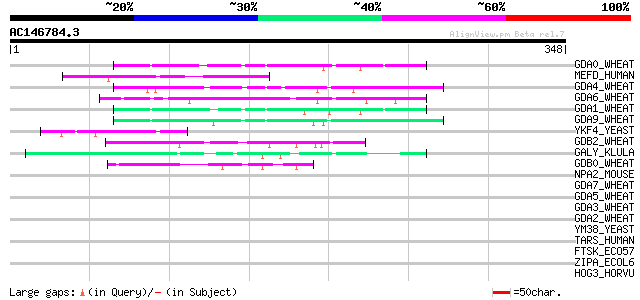
Score E
Sequences producing significant alignments: (bits) Value
GDA0_WHEAT (P02863) Alpha/beta-gliadin precursor (Prolamin) 56 1e-07
MEFD_HUMAN (Q14814) Myocyte-specific enhancer factor 2D 50 8e-06
GDA4_WHEAT (P04724) Alpha/beta-gliadin A-IV precursor (Prolamin) 48 3e-05
GDA6_WHEAT (P04726) Alpha/beta-gliadin clone PW1215 precursor (P... 47 9e-05
GDA1_WHEAT (P04721) Alpha/beta-gliadin A-I precursor (Prolamin) 47 9e-05
GDA9_WHEAT (P18573) Alpha/beta-gliadin MM1 precursor (Prolamin) 46 2e-04
YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4... 45 2e-04
GDB2_WHEAT (P08453) Gamma-gliadin precursor 45 3e-04
GALY_KLULA (P32257) Transcription regulatory protein GAL11 44 6e-04
GDB0_WHEAT (P08079) Gamma-gliadin precursor (Fragment) 44 8e-04
NPA2_MOUSE (P97460) Neuronal PAS domain protein 2 (Neuronal PAS2) 42 0.002
GDA7_WHEAT (P04727) Alpha/beta-gliadin clone PW8142 precursor (P... 42 0.002
GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin) 41 0.004
GDA3_WHEAT (P04723) Alpha/beta-gliadin A-III precursor (Prolamin) 41 0.005
GDA2_WHEAT (P04722) Alpha/beta-gliadin A-II precursor (Prolamin) 40 0.007
YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2 i... 39 0.015
TARS_HUMAN (Q7Z7G0) Target of Nesh-SH3 precursor (Tarsh) (Nesh b... 39 0.015
FTSK_ECO57 (Q8X5H9) DNA translocase ftsK 39 0.015
ZIPA_ECOL6 (Q8FFC0) Cell division protein zipA 39 0.019
HOG3_HORVU (P80198) Gamma-hordein 3 39 0.025
>GDA0_WHEAT (P02863) Alpha/beta-gliadin precursor (Prolamin)
Length = 286
Score = 56.2 bits (134), Expect = 1e-07
Identities = 60/213 (28%), Positives = 87/213 (40%), Gaps = 28/213 (13%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + LP L +
Sbjct: 24 PVPQLQPQNPSQQLPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQLPYL----QL 78
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIK 185
Q P P++P S P +RP + Q P + Q P+ ++ Q+ + Q I
Sbjct: 79 QPFPQPQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQQIL 135
Query: 186 VLLQQQHLAP--------HSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPDSH 228
+ QQ L P H++A R + V+Q YQ P Q Q H
Sbjct: 136 QQILQQQLIPCMDVVLQQHNIAHGR--SQVLQQSTYQLLQELCCQHLWQIPEQSQCQAIH 193
Query: 229 FVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
V + I++ + QP Q QQP QQ P
Sbjct: 194 NVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 225
Score = 37.0 bits (84), Expect = 0.074
Identities = 49/205 (23%), Positives = 76/205 (36%), Gaps = 28/205 (13%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +L +L+
Sbjct: 83 QPQLPYSQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQ-QQQQQQQQQILQQILQ 140
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIENNVWSF 179
+ L+ P ++ V Q H Q + +Q+L ++W
Sbjct: 141 QQLI-----------------PCMDVVLQQHNIAHGRSQVLQQSTYQLLQELCCQHLWQI 183
Query: 180 DDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP---DSHFVTTILPI 236
+Q Q H H++ + Q P Q F+ QQYP S + P
Sbjct: 184 PEQS---QCQAIHNVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYPLGQGSFRPSQQNPQ 239
Query: 237 MNVVRNPGYQPQFQQYQQQPRQQAP 261
P PQF++ + Q P
Sbjct: 240 AQGSVQPQQLPQFEEIRNLALQTLP 264
>MEFD_HUMAN (Q14814) Myocyte-specific enhancer factor 2D
Length = 521
Score = 50.1 bits (118), Expect = 8e-06
Identities = 38/137 (27%), Positives = 58/137 (41%), Gaps = 20/137 (14%)
Query: 34 TTLRAEVEKLASLVSSLMATNDPPLVQ-------QRPQSPYQPMCPQKPRQQAPRQSIPQ 86
T + + ++ +SSL A + P + Q+PQ P QP PQ P+QQ P+ PQ
Sbjct: 331 TAYNTDYQLTSAELSSLPAFSSPGGLSLGNVTAWQQPQQPQQPQQPQPPQQQPPQPQQPQ 390
Query: 87 NQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLN 146
Q PQ+ PQ Q+ S P+ + NL+ PLP V +L P ++
Sbjct: 391 PQQPQQ--PQQPPQQQSHLVPVSL-----------SNLIPGSPLPHVGAALTVTTHPHIS 437
Query: 147 CVFHQGAPGHDTEQCYP 163
+P + P
Sbjct: 438 IKSEPVSPSRERSPAPP 454
>GDA4_WHEAT (P04724) Alpha/beta-gliadin A-IV precursor (Prolamin)
Length = 297
Score = 48.1 bits (113), Expect = 3e-05
Identities = 59/222 (26%), Positives = 91/222 (40%), Gaps = 25/222 (11%)
Query: 66 PYQPMCPQKPRQQAPRQSIP---QNQVP--QKFIPQNQVQKASQCDPIPMKYADLLPILL 120
P + PQ P QQ P++ +P Q Q P Q+ P Q Q P Y L P
Sbjct: 24 PVPQLQPQNPSQQQPQKQVPLVQQQQFPGQQQPFPPQQPYPQQQPFPSQQPYMQLQPFPQ 83
Query: 121 KKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD 180
+ P P++P P +RP + + Q P + Q P+ ++ Q+ +
Sbjct: 84 PQ---LPYPQPQLPYPQPQPFRPQQS--YPQPQPQYSQPQ-QPISQQQQQ--QQQQQQQQ 135
Query: 181 DQDIKVLLQQQ-------HLAPHSVAAVRPITNVVQDPGYQ--PQFRPSQ-QQYPDSHFV 230
Q ++ +LQQQ L HS+A + V+Q YQ QF Q Q P+
Sbjct: 136 QQILQQILQQQLIPCRDVVLQQHSIA--HGSSQVLQQSTYQLVQQFCCQQLWQIPEQSRC 193
Query: 231 TTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKY 272
I +++ + Q Q QQ QQQ +Q ++ F +Y
Sbjct: 194 QAIHNVVHAIILHQQQQQQQQQQQQQQQPLSQVCFQQSQQQY 235
Score = 40.8 bits (94), Expect = 0.005
Identities = 52/214 (24%), Positives = 78/214 (36%), Gaps = 55/214 (25%)
Query: 56 PPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADL 115
P L +PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +
Sbjct: 84 PQLPYPQPQLPYPQPQPFRPQQSYP-QPQPQYSQPQQPISQQQQQQQQQ----QQQQQQI 138
Query: 116 LPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIE-- 173
L +L++ L+ P + V Q + H + Q L++ +L++
Sbjct: 139 LQQILQQQLI-----------------PCRDVVLQQHSIAHGSSQV--LQQSTYQLVQQF 179
Query: 174 --NNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQ------FRPSQQQYP 225
+W +Q Q H H++ + Q Q Q F+ SQQQYP
Sbjct: 180 CCQQLWQIPEQS---RCQAIHNVVHAIILHQQQQQQQQQQQQQQQPLSQVCFQQSQQQYP 236
Query: 226 DSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQ 259
Q FQ QQ P+ Q
Sbjct: 237 SG------------------QGSFQPSQQNPQAQ 252
>GDA6_WHEAT (P04726) Alpha/beta-gliadin clone PW1215 precursor
(Prolamin)
Length = 296
Score = 46.6 bits (109), Expect = 9e-05
Identities = 61/221 (27%), Positives = 90/221 (40%), Gaps = 27/221 (12%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMK--YAD 114
P+ Q +PQ+P QP PQ +Q P Q Q+F PQ Q Q P P + Y
Sbjct: 24 PVPQPQPQNPSQPQ-PQGQVPLVQQQQFPGQQ--QQFPPQ---QPYPQPQPFPSQQPYLQ 77
Query: 115 LLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIEN 174
L P + LP P+ P P P + Q +Q +++ Q+ +
Sbjct: 78 LQPFPQPQPFPPQLPYPQPPPFSPQQPYPQPQPQYPQPQQPISQQQAQQQQQQQQQQQQQ 137
Query: 175 NVWSFDDQDIKVLLQQQ-------HLAPHSVAAVRPITNVVQDPGYQP-QFRPSQQ--QY 224
Q ++ +LQQQ L H++A R + V+Q YQP Q QQ Q
Sbjct: 138 QQ---QQQILQQILQQQLIPCRDVVLQQHNIAHAR--SQVLQQSTYQPLQQLCCQQLWQI 192
Query: 225 PDSHFVTTILPIMNVV----RNPGYQPQFQQYQQQPRQQAP 261
P+ I +++ + + QP Q QQP+QQ P
Sbjct: 193 PEQSRCQAIHNVVHAIILHQQQRQQQPSSQVSLQQPQQQYP 233
Score = 38.1 bits (87), Expect = 0.033
Identities = 49/212 (23%), Positives = 74/212 (34%), Gaps = 34/212 (16%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLL 116
P Q P SP QP +P+ P+Q I Q Q Q+ Q Q Q+ Q +L
Sbjct: 92 PYPQPPPFSPQQPYPQPQPQYPQPQQPISQQQAQQQQQQQQQQQQQQQ-------QQQIL 144
Query: 117 PILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIEN 174
+L++ L+ P + V Q H Q + +Q+L
Sbjct: 145 QQILQQQLI-----------------PCRDVVLQQHNIAHARSQVLQQSTYQPLQQLCCQ 187
Query: 175 NVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFV---T 231
+W +Q Q H H++ + Q P Q + QQQYP +
Sbjct: 188 QLWQIPEQS---RCQAIHNVVHAIILHQQQRQ--QQPSSQVSLQQPQQQYPSGQGFFQPS 242
Query: 232 TILPIMNVVRNPGYQPQFQQYQQQPRQQAPRI 263
P P PQF++ + Q PR+
Sbjct: 243 QQNPQAQGSVQPQQLPQFEEIRNLALQTLPRM 274
Score = 32.0 bits (71), Expect = 2.4
Identities = 29/100 (29%), Positives = 40/100 (40%), Gaps = 17/100 (17%)
Query: 205 TNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIK 264
T V+ P QPQ + Q P +P++ + PG Q QF Q P+ Q
Sbjct: 18 TTAVRVPVPQPQPQNPSQPQPQGQ-----VPLVQQQQFPGQQQQFPPQQPYPQPQ----- 67
Query: 265 FDPIPMKYGELFPYLLERNLVQTRP-PPPIPKKLPARWRP 303
P P PYL + Q +P PP +P P + P
Sbjct: 68 --PFP----SQQPYLQLQPFPQPQPFPPQLPYPQPPPFSP 101
>GDA1_WHEAT (P04721) Alpha/beta-gliadin A-I precursor (Prolamin)
Length = 262
Score = 46.6 bits (109), Expect = 9e-05
Identities = 57/212 (26%), Positives = 85/212 (39%), Gaps = 25/212 (11%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F+ Q Q Q P P + P L + +
Sbjct: 24 PVPQLQPQNPSQQQPQEQVPLVQ-QQQFLGQQQPFPPQQPYPQPQPFPSQQPYLQLQPFL 82
Query: 126 QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQD-- 183
Q P++P S P +RP + Q P + Q P+ ++ Q+ + Q
Sbjct: 83 Q----PQLPYSQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQQQQQQQQQQQQQQQI 135
Query: 184 IKVLLQQQHLAPHSVA-----AVRPITNVVQDPGYQPQFR---------PSQQQYPDSHF 229
I+ +LQQQ + V V + V+Q YQ P Q Q H
Sbjct: 136 IQQILQQQLIPCMDVVLQQHNIVHGKSQVLQQSTYQLLQELCCQHLWQIPEQSQCQAIHN 195
Query: 230 VTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
V + I++ + QP Q QQP QQ P
Sbjct: 196 VVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 226
Score = 38.1 bits (87), Expect = 0.033
Identities = 40/166 (24%), Positives = 64/166 (38%), Gaps = 24/166 (14%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + ++ +L+
Sbjct: 83 QPQLPYSQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQQQQQQQQQQQIIQQILQ 141
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQKLIENNVWSF 179
+ L+ P ++ V Q H Q + +Q+L ++W
Sbjct: 142 QQLI-----------------PCMDVVLQQHNIVHGKSQVLQQSTYQLLQELCCQHLWQI 184
Query: 180 DDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYP 225
+Q Q H H++ + Q P Q F+ QQYP
Sbjct: 185 PEQS---QCQAIHNVVHAI-ILHQQQKQQQQPSSQVSFQQPLQQYP 226
>GDA9_WHEAT (P18573) Alpha/beta-gliadin MM1 precursor (Prolamin)
Length = 307
Score = 45.8 bits (107), Expect = 2e-04
Identities = 59/228 (25%), Positives = 87/228 (37%), Gaps = 27/228 (11%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLV 125
P + PQ P QQ P++ +P Q Q+F Q Q Q P P + P L +
Sbjct: 24 PVPQLQPQNPSQQQPQEQVPLVQ-QQQFPGQQQPFPPQQPYPQPQPFPSQQPYLQLQPFP 82
Query: 126 Q----------TLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENN 175
Q P P++P P +RP + Q P + Q P+ ++ Q+ +
Sbjct: 83 QPQLPYPQPQLPYPQPQLPYPQPQPFRPQQ--PYPQSQPQYSQPQ-QPISQQQQQQQQQQ 139
Query: 176 VWSFDDQDIKVLLQ---QQHLAP--------HSVAAVRPITNVVQDPGYQPQFRPSQQQY 224
Q + +LQ QQ L P HS+A + V+Q YQ + QQ
Sbjct: 140 QQKQQQQQQQQILQQILQQQLIPCRDVVLQQHSIA--YGSSQVLQQSTYQLVQQLCCQQL 197
Query: 225 PDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIPMKY 272
+ I NVV Q QQ QQQ +Q ++ F +Y
Sbjct: 198 WQIPEQSRCQAIHNVVHAIILHQQQQQQQQQQQQPLSQVSFQQPQQQY 245
Score = 38.9 bits (89), Expect = 0.019
Identities = 52/213 (24%), Positives = 78/213 (36%), Gaps = 50/213 (23%)
Query: 56 PPLVQQRPQSPY---QPMCPQKPRQQA-PRQSIPQNQVPQKFIPQNQVQKASQCDPIPMK 111
P L +PQ PY QP PQ+P Q+ P+ S PQ + Q+ Q Q Q+ Q +
Sbjct: 91 PQLPYPQPQLPYPQPQPFRPQQPYPQSQPQYSQPQQPISQQ---QQQQQQQQQQKQQQQQ 147
Query: 112 YADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLK--EEVQ 169
+L +L++ L+ P + V Q + + + Q + VQ
Sbjct: 148 QQQILQQILQQQLI-----------------PCRDVVLQQHSIAYGSSQVLQQSTYQLVQ 190
Query: 170 KLIENNVWSFDDQDIKVLLQQQHLAPHSV---AAVRPITNVVQDPGYQPQFRPSQQQYPD 226
+L +W +Q Q H H++ + Q P Q F+ QQQYP
Sbjct: 191 QLCCQQLWQIPEQS---RCQAIHNVVHAIILHQQQQQQQQQQQQPLSQVSFQQPQQQYPS 247
Query: 227 SHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQ 259
Q FQ QQ P+ Q
Sbjct: 248 G------------------QGSFQPSQQNPQAQ 262
>YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4
intergenic region
Length = 738
Score = 45.4 bits (106), Expect = 2e-04
Identities = 33/97 (34%), Positives = 51/97 (52%), Gaps = 8/97 (8%)
Query: 20 DQLEQEVQELRG---EVTTLRAEVEKLASLVSSLMA--TNDPPLVQQRPQSPYQPMCPQK 74
++LEQE + EVT + +VE +++++S N P QQ+ Q P QP PQ+
Sbjct: 340 EELEQEQDNVAAPEEEVTVVEEKVE-ISAVISEPPEDQANTVPQPQQQSQQPQQPQQPQQ 398
Query: 75 PRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMK 111
P+Q Q Q Q PQ+ PQ Q+Q+ Q P++
Sbjct: 399 PQQPQQPQQQQQPQQPQQ--PQQQLQQQQQQQQQPVQ 433
Score = 32.3 bits (72), Expect = 1.8
Identities = 18/44 (40%), Positives = 24/44 (53%), Gaps = 2/44 (4%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQ 100
P Q+PQ P Q PQ+P+Q P+Q + Q Q Q+ Q Q Q
Sbjct: 396 PQQPQQPQQPQQQQQPQQPQQ--PQQQLQQQQQQQQQPVQAQAQ 437
Score = 31.2 bits (69), Expect = 4.0
Identities = 33/132 (25%), Positives = 49/132 (37%), Gaps = 17/132 (12%)
Query: 61 QRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQ----KFIPQNQVQKASQCDPIPMKYADLL 116
Q+PQ P Q + Q+ +QQ P Q+ Q Q Q + Q Q Q A Q + +Y
Sbjct: 412 QQPQQPQQQLQQQQQQQQQPVQAQAQAQEEQLSQNYYTQQQQQQYAQQQHQLQQQYLSQQ 471
Query: 117 PILLKKNLVQTLPLPRVPNS--LPPWYRPDLNCVFHQ-----------GAPGHDTEQCYP 163
++ P + P S P + N Q PG Q Y
Sbjct: 472 QQYAQQQQQHPQPQSQQPQSQQSPQSQKQGNNVAAQQYYMYQNQFPGYSYPGMFDSQGYA 531
Query: 164 LKEEVQKLIENN 175
++ Q+L +NN
Sbjct: 532 YGQQYQQLAQNN 543
Score = 30.0 bits (66), Expect = 9.0
Identities = 25/92 (27%), Positives = 37/92 (40%), Gaps = 4/92 (4%)
Query: 15 QTGNMDQLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDPPLVQQR--PQSPYQPMCP 72
Q +Q +E+++ + V EV + V ++PP Q PQ Q P
Sbjct: 331 QVKEEEQTAEELEQEQDNVAAPEEEVTVVEEKVEISAVISEPPEDQANTVPQPQQQSQQP 390
Query: 73 QKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQ 104
Q+P Q P+Q Q Q+ PQ Q Q
Sbjct: 391 QQP--QQPQQPQQPQQPQQQQQPQQPQQPQQQ 420
>GDB2_WHEAT (P08453) Gamma-gliadin precursor
Length = 327
Score = 45.1 bits (105), Expect = 3e-04
Identities = 54/177 (30%), Positives = 74/177 (41%), Gaps = 23/177 (12%)
Query: 61 QRPQSPY-QPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQC----DPIPMKYADL 115
Q+PQ P+ Q PQ+P Q P+Q PQ Q PQ+ PQ Q Q P P
Sbjct: 86 QQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQLQQPQ 145
Query: 116 LPILLKKNLVQTLPLPRVP-NSLPPWYRPDLNCVFHQGAPGHDTEQC--YPLKEEVQKLI 172
P + Q LP P+ P S P RP F Q + C L++ +
Sbjct: 146 QPFPQPQ---QQLPQPQQPQQSFPQQQRP-----FIQPSLQQQLNPCKNILLQQSKPASL 197
Query: 173 ENNVWS--FDDQDIKVLLQQ--QHLA--PHSVAAVRPITNVVQDPGYQPQFRPSQQQ 223
+++WS + D +V+ QQ Q LA P + I +VV Q Q + QQQ
Sbjct: 198 VSSLWSIIWPQSDCQVMRQQCCQQLAQIPQQLQCA-AIHSVVHSIIMQQQQQQQQQQ 253
>GALY_KLULA (P32257) Transcription regulatory protein GAL11
Length = 1034
Score = 43.9 bits (102), Expect = 6e-04
Identities = 58/259 (22%), Positives = 102/259 (38%), Gaps = 45/259 (17%)
Query: 11 QTLVQTGNMDQLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDPPLVQQRPQSPYQP- 69
Q Q Q +Q+ Q + + + + + + +++ M N +Q+ Q +
Sbjct: 94 QAQAQAVQRQQQQQQQQHHMQQQGSGQQQANGIPANINAQMFLNQQAQARQQAQRQIRST 153
Query: 70 MCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLP 129
+ Q+ +Q P+Q PQ Q P PQ +Q+ Q +K ++ LL+K
Sbjct: 154 LTGQQQQQPMPQQPQPQQQQPNMMRPQLTLQQQQQL-ANELKVTEIPRELLQK------- 205
Query: 130 LPRVPNSLPPWYRPDLNCVFHQGAPGHD----TEQCYPLKEEV---QKLIENNVWSFDDQ 182
+P +P + W P + + P + T++ Y L +++ KL ++NV
Sbjct: 206 IPNLPPGITTW--PQITFWAQKNRPSQNDLIITKKVYQLHQQLLNKSKLQQSNV-----N 258
Query: 183 DIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRN 242
+ +QQ + R N+ Q P Q Q R QQQ
Sbjct: 259 RMNTSVQQPGMGVQQSPNQR--MNMNQVPQQQQQVRQQQQQQ------------------ 298
Query: 243 PGYQPQFQQYQQQPRQQAP 261
Q Q QQ++QQP+ Q P
Sbjct: 299 --QQQQQQQHKQQPQHQEP 315
Score = 30.4 bits (67), Expect = 6.9
Identities = 25/84 (29%), Positives = 34/84 (39%), Gaps = 1/84 (1%)
Query: 189 QQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPG-YQP 247
QQQ + I V Q P QPQ +P Q Q S + V+ N G +P
Sbjct: 400 QQQQQQQQQQQRQQQIPQVQQAPLQQPQVQPPQSQQAQSRRQAMGMKPTPVIPNAGVMKP 459
Query: 248 QFQQYQQQPRQQAPRIKFDPIPMK 271
Q QQ Q +Q + + P P +
Sbjct: 460 QPQQTPQNIQQPMMQQQSSPPPQQ 483
>GDB0_WHEAT (P08079) Gamma-gliadin precursor (Fragment)
Length = 251
Score = 43.5 bits (101), Expect = 8e-04
Identities = 38/138 (27%), Positives = 58/138 (41%), Gaps = 36/138 (26%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ P+ P PQ+P Q P+Q PQ Q PQ+ PQ+Q + P
Sbjct: 79 QPQQPF-PQQPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQQFSQP-------------- 123
Query: 122 KNLVQTLPLPR-----VPNSLPPWYRPDLNCVFHQGAPGHD--TEQCYPLKEEVQKLIEN 174
Q P P+ P PP+ +P L Q P + +QC P+ + +
Sbjct: 124 ---QQQFPQPQQPQQSFPQQQPPFIQPSLQ---QQVNPCKNFLLQQCKPVS------LVS 171
Query: 175 NVWS--FDDQDIKVLLQQ 190
++WS + D +V+ QQ
Sbjct: 172 SLWSMIWPQSDCQVMRQQ 189
Score = 32.3 bits (72), Expect = 1.8
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 2/45 (4%)
Query: 61 QRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQC 105
Q+PQ + Q P+ Q P+QS PQ Q P FI + Q+ + C
Sbjct: 114 QQPQQQFSQPQQQFPQPQQPQQSFPQQQPP--FIQPSLQQQVNPC 156
>NPA2_MOUSE (P97460) Neuronal PAS domain protein 2 (Neuronal PAS2)
Length = 816
Score = 42.4 bits (98), Expect = 0.002
Identities = 80/322 (24%), Positives = 119/322 (36%), Gaps = 61/322 (18%)
Query: 20 DQLEQEVQELRGEVTTLRAEVEKL------------------ASLVSSLMATNDPPLVQQ 61
DQLEQ + L+ + + E+ K+ ++ S +T P QQ
Sbjct: 511 DQLEQRTRILQANIRWQQEELHKIQEQLCLVQDSNVQMFLQQPAVSLSFSSTQRPAAQQQ 570
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVP-QKFIPQNQVQKASQCDPIPMKYADLLPILL 120
Q P P PQ + I QV Q + ++ V A P PM+ + LLP
Sbjct: 571 LQQRPAAPSQPQLVVNTPLQGQITSTQVTNQHLLRESNVISAQ--GPKPMRSSQLLPASG 628
Query: 121 KKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD 180
+ LP +S P L+ P D QC P +
Sbjct: 629 R----SLSSLPSQFSSTASVLPPGLSLTTIAPTP-QDDSQCQPSPD-----------FGH 672
Query: 181 DQDIKVLLQQ--QHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQY--PDSH-----FVT 231
D+ +++LL Q Q + P S A +P + V G Q ++ SQ + PDSH T
Sbjct: 673 DRQLRLLLSQPIQPMMPGSCDARQP--SEVSRTGRQVKYAQSQVMFPSPDSHPTNSSAST 730
Query: 232 TILPIMNVVRNPGY--------QPQFQQYQQQPRQQAPRI----KFDPIPMKYGELFPYL 279
+L + V +P + QP Q Q P QAP + D + + P
Sbjct: 731 PVLLMGQAVLHPSFPASRPSPLQPAQAQQQPPPYLQAPTSLHSEQPDSLLLSTFSQQPGT 790
Query: 280 LERNLVQTRPP-PPIPKKLPAR 300
L Q+ PP PP P + +R
Sbjct: 791 LGYAATQSTPPQPPRPSRRVSR 812
>GDA7_WHEAT (P04727) Alpha/beta-gliadin clone PW8142 precursor
(Prolamin)
Length = 313
Score = 42.4 bits (98), Expect = 0.002
Identities = 58/232 (25%), Positives = 95/232 (40%), Gaps = 29/232 (12%)
Query: 46 LVSSLMATNDP-----PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVP--QKFIPQNQ 98
L+ +L+AT P+ Q +P++P Q Q+P++Q P + Q Q P Q+ P Q
Sbjct: 5 LILALVATTATTAVRVPVPQLQPKNPSQ----QQPQEQVP--LVQQQQFPGQQQQFPPQQ 58
Query: 99 VQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSLPPW---------YRPDLNCVF 149
Q P Y L P + + LP P+ P S PP Y +
Sbjct: 59 PYPQPQPFPSQQPYLQLQPFPQPQPFLPQLPYPQ-PQSFPPQQPYPQQRPKYLQPQQPIS 117
Query: 150 HQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAPHSVAAVRPIT-NVV 208
Q A +Q +++ Q++++ + V+LQQ ++A S ++ T ++
Sbjct: 118 QQQAQQQQQQQQQQQQQQQQQILQQILQQQLIPCRDVVLQQHNIAHASSQVLQQSTYQLL 177
Query: 209 QDPGYQPQFR-PSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQ 259
Q Q + P Q + H V + ++ Q Q QQ QQQ QQ
Sbjct: 178 QQLCCQQLLQIPEQSRCQAIHNVVHAI----IMHQQEQQQQLQQQQQQQLQQ 225
>GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin)
Length = 319
Score = 41.2 bits (95), Expect = 0.004
Identities = 61/222 (27%), Positives = 89/222 (39%), Gaps = 36/222 (16%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVP--QKFIPQNQVQKASQCDPIPMKYAD 114
P+ Q +PQ+P Q Q+P++Q P + Q Q P Q+ P Q Q P Y
Sbjct: 24 PVPQLQPQNPSQ----QQPQEQVP--LVQQQQFPGQQQQFPPQQPYPQPQPFPSQQPYLQ 77
Query: 115 LLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIEN 174
L P + LP P+ P S PP +P + Q P + Q +++ Q+ +
Sbjct: 78 LQPFPQPQPFPPQLPYPQ-PQSFPP-QQP-----YPQQQPQYLQPQQPISQQQAQQQQQQ 130
Query: 175 NVWSFDDQDIKVLLQQQHLAP--------HSVAAVRPITNVVQDPGYQPQFR-------- 218
Q I + QQ L P H++A + V+Q YQ +
Sbjct: 131 QQQQQQQQQILQQILQQQLIPCRDVVLQQHNIA--HASSQVLQQSTYQLLQQLCCQQLLQ 188
Query: 219 -PSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQ 259
P Q Q H V + IM+ + Q Q QQ QQQ +QQ
Sbjct: 189 IPEQSQCQAIHNVAHAI-IMH-QQQQQQQEQKQQLQQQQQQQ 228
Score = 39.3 bits (90), Expect = 0.015
Identities = 64/247 (25%), Positives = 94/247 (37%), Gaps = 53/247 (21%)
Query: 56 PPLVQQRPQS--PYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYA 113
P L +PQS P QP Q+P+ P+Q I Q Q Q+ Q Q Q+ Q
Sbjct: 89 PQLPYPQPQSFPPQQPYPQQQPQYLQPQQPISQQQAQQQQQQQQQQQQQQQ--------- 139
Query: 114 DLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIE 173
+L +L++ L+ P + V Q H + Q L++ +L++
Sbjct: 140 -ILQQILQQQLI-----------------PCRDVVLQQHNIAHASSQV--LQQSTYQLLQ 179
Query: 174 NNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTI 233
Q +++ Q Q A H+VA + Q Q Q QQQ
Sbjct: 180 QLCC---QQLLQIPEQSQCQAIHNVAHAIIMHQQQQQQQEQKQQLQQQQQQQQQ------ 230
Query: 234 LPIMNVVRNPGYQPQFQQYQQQPRQQAP--RIKFDPI---PMKYGELFPYLLE-----RN 283
+ + QP Q QQP+QQ P ++ F P P G + P L RN
Sbjct: 231 --LQQQQQQQQQQPSSQVSFQQPQQQYPSSQVSFQPSQLNPQAQGSVQPQQLPQFAEIRN 288
Query: 284 L-VQTRP 289
L +QT P
Sbjct: 289 LALQTLP 295
Score = 33.9 bits (76), Expect = 0.62
Identities = 36/113 (31%), Positives = 48/113 (41%), Gaps = 20/113 (17%)
Query: 193 LAPHSVAAVRPITNV-VQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQ 251
LA ++ A T V V P QPQ PSQQQ + +P++ + PG Q QF
Sbjct: 7 LALLAIVATTATTAVRVPVPQLQPQ-NPSQQQPQEQ------VPLVQQQQFPGQQQQFPP 59
Query: 252 YQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRP-PPPIPKKLPARWRP 303
Q P+ Q P P PYL + Q +P PP +P P + P
Sbjct: 60 QQPYPQPQ-------PFP----SQQPYLQLQPFPQPQPFPPQLPYPQPQSFPP 101
>GDA3_WHEAT (P04723) Alpha/beta-gliadin A-III precursor (Prolamin)
Length = 282
Score = 40.8 bits (94), Expect = 0.005
Identities = 51/214 (23%), Positives = 81/214 (37%), Gaps = 36/214 (16%)
Query: 66 PYQPMCPQKPRQQAPRQSIPQNQVPQKF------IPQNQVQKASQCDPIPMKYADLLPIL 119
P + PQ P QQ P++ +P Q Q+F P Q Q P Y P
Sbjct: 24 PVPQLQPQNPSQQQPQEQVPLMQQQQQFPGQQEQFPPQQPYPHQQPFPSQQPYPQPQPFP 83
Query: 120 LKKNLVQTLPLP---RVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNV 176
+ QT P P P P + +P Q +Q ++ +Q++++ +
Sbjct: 84 PQLPYPQTQPFPPQQPYPQPQPQYPQP------QQPISQQQAQQQQQQQQTLQQILQQQL 137
Query: 177 WSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFR---------PSQQQYPDS 227
D V+LQQ ++A S + V+Q YQ + P Q +
Sbjct: 138 IPCRD----VVLQQHNIAHAS-------SQVLQQSSYQQLQQLCCQQLFQIPEQSRCQAI 186
Query: 228 HFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAP 261
H V + I++ + QP Q QQP++Q P
Sbjct: 187 HNVVHAI-ILHHHQQQQQQPSSQVSYQQPQEQYP 219
Score = 32.3 bits (72), Expect = 1.8
Identities = 18/55 (32%), Positives = 26/55 (46%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMK 111
P Q +P P QP +P+ P+Q I Q Q Q+ Q +Q+ Q IP +
Sbjct: 87 PYPQTQPFPPQQPYPQPQPQYPQPQQPISQQQAQQQQQQQQTLQQILQQQLIPCR 141
Score = 31.6 bits (70), Expect = 3.1
Identities = 41/150 (27%), Positives = 63/150 (41%), Gaps = 27/150 (18%)
Query: 193 LAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRN-PGYQPQFQQ 251
+A + +AVR V P QPQ PSQQQ + +P+M + PG Q QF
Sbjct: 13 VATTATSAVR-----VPVPQLQPQ-NPSQQQPQEQ------VPLMQQQQQFPGQQEQFP- 59
Query: 252 YQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPP---PIPKKLPARWRPDLFCV 308
P+Q P + P Y + P+ + QT+P P P P+ P +P
Sbjct: 60 ----PQQPYPHQQPFPSQQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPIS 115
Query: 309 FHQGAQGHDVERCFSLKIEVQKLIEDDLIP 338
Q Q ++ +Q++++ LIP
Sbjct: 116 QQQAQQQQQQQQ------TLQQILQQQLIP 139
>GDA2_WHEAT (P04722) Alpha/beta-gliadin A-II precursor (Prolamin)
Length = 291
Score = 40.4 bits (93), Expect = 0.007
Identities = 57/217 (26%), Positives = 90/217 (41%), Gaps = 35/217 (16%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLL 116
P+ Q + Q+P Q Q+P++Q P Q Q Q+ P Q Q P Y L
Sbjct: 24 PVPQLQLQNPSQ----QQPQEQVPLVQEQQFQGQQQPFPPQQPYPQPQPFPSQQPYLQLQ 79
Query: 117 PILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNV 176
P P P++P P +RP + Q P + Q P+ ++ Q+ +
Sbjct: 80 P----------FPQPQLPYPQPQPFRPQQ--PYPQPQPQYSQPQ-QPISQQQQQ--QQQQ 124
Query: 177 WSFDDQDIKVLLQQQ-------HLAPHSVAAVRPITNVVQDPGYQ-PQFRPSQQ--QYPD 226
Q ++ +LQQQ L H++A + V+Q+ YQ Q QQ Q P+
Sbjct: 125 QQQQQQILQQILQQQLIPCRDVVLQQHNIA--HGSSQVLQESTYQLVQQLCCQQLWQIPE 182
Query: 227 SHFVTTILPIMNVV----RNPGYQPQFQQYQQQPRQQ 259
I +++ + ++ +Q Q QQ QQQP Q
Sbjct: 183 QSRCQAIHNVVHAIILHQQHHHHQQQQQQQQQQPLSQ 219
Score = 37.7 bits (86), Expect = 0.043
Identities = 53/221 (23%), Positives = 79/221 (34%), Gaps = 47/221 (21%)
Query: 62 RPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLK 121
+PQ PY P +P+Q P Q PQ PQ+ I Q Q Q+ Q + +L +L+
Sbjct: 83 QPQLPYPQPQPFRPQQPYP-QPQPQYSQPQQPISQQQQQQQQQ----QQQQQQILQQILQ 137
Query: 122 KNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEE----VQKLIENNVW 177
+ L+ P + V Q H + Q L+E VQ+L +W
Sbjct: 138 QQLI-----------------PCRDVVLQQHNIAHGSSQV--LQESTYQLVQQLCCQQLW 178
Query: 178 SFDDQD---------IKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSH 228
+Q ++L QQH + Q P Q F+ QQQYP
Sbjct: 179 QIPEQSRCQAIHNVVHAIILHQQHHHHQQQQQQQQ-----QQPLSQVSFQQPQQQYPSGQ 233
Query: 229 FVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIP 269
+NP Q FQ Q ++ + +P
Sbjct: 234 GF-----FQPSQQNPQAQGSFQPQQLPQFEEIRNLALQTLP 269
>YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2
intergenic region
Length = 758
Score = 39.3 bits (90), Expect = 0.015
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 10/87 (11%)
Query: 208 VQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDP 267
+Q P +QPQ +P QQQ + Q Q QQ QQQ +QQ + + P
Sbjct: 282 IQQPQHQPQHQPQQQQQQQQQ----------QQQQQQQQQQQQQQQQQQQQQHQQQQQTP 331
Query: 268 IPMKYGELFPYLLERNLVQTRPPPPIP 294
P+ ++ P++ N T P +P
Sbjct: 332 YPIVNPQMVPHIPSENSHSTGLMPSVP 358
Score = 38.9 bits (89), Expect = 0.019
Identities = 32/140 (22%), Positives = 60/140 (42%), Gaps = 1/140 (0%)
Query: 60 QQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPIL 119
Q + Q +QP Q+ +QQ +Q Q Q Q+ Q Q + Q P P+ ++P +
Sbjct: 284 QPQHQPQHQPQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQTPYPIVNPQMVPHI 343
Query: 120 LKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSF 179
+N T +P VP + + + +F + + + + ++ F
Sbjct: 344 PSENSHSTGLMPSVPPTNQQFNAQTQSSMFSDQQRFFQYQLHHQNQGQAPSFQQSQSGRF 403
Query: 180 DDQD-IKVLLQQQHLAPHSV 198
DD + +K+ QQQ L +S+
Sbjct: 404 DDMNAMKMFFQQQALQQNSL 423
>TARS_HUMAN (Q7Z7G0) Target of Nesh-SH3 precursor (Tarsh) (Nesh
binding protein) (NeshBP)
Length = 1075
Score = 39.3 bits (90), Expect = 0.015
Identities = 53/217 (24%), Positives = 81/217 (36%), Gaps = 22/217 (10%)
Query: 53 TNDPPLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKY 112
T+ P +Q P++P +P + +PR IPQ Q P +PQ K + Y
Sbjct: 528 THTKPAPKQTPRAPPKP-------KTSPRPRIPQTQ-PVPKVPQRVTAKPKTSPSPEVSY 579
Query: 113 ADLLPILLKKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLI 172
P K L+ P P V S P +G P P +EE+Q +
Sbjct: 580 TTPAP---KDVLLPHKPYPEVSQSEPAPLET-------RGIPFIPMISPSPSQEELQTTL 629
Query: 173 ENNVWSFDDQDIKVLLQQQHLAPHSVAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTT 232
E S + + + LA + A R T V +P RP +Q P
Sbjct: 630 EETDQSTQEPFTTKIPRTTELAKTTQAPHRFYTTVRPRTSDKPHIRPGVKQAPRPSGADR 689
Query: 233 ILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPIP 269
+ + + +P +P ++ PR PR K P+P
Sbjct: 690 NVSVDST--HPTKKPGTRRPPLPPRPTHPRRK--PLP 722
>FTSK_ECO57 (Q8X5H9) DNA translocase ftsK
Length = 1342
Score = 39.3 bits (90), Expect = 0.015
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 5/63 (7%)
Query: 46 LVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQQAPRQSI---PQNQVPQKFI-PQNQVQK 101
+V + P QQ+ Q P QP+ PQ P+ Q P+Q + PQ Q PQ+ + PQ Q Q+
Sbjct: 735 IVEPVQQPQQPVAPQQQYQQPQQPVAPQ-PQYQQPQQQVAPQPQYQQPQQPVAPQQQYQQ 793
Query: 102 ASQ 104
Q
Sbjct: 794 PQQ 796
Score = 37.0 bits (84), Expect = 0.074
Identities = 31/88 (35%), Positives = 43/88 (48%), Gaps = 13/88 (14%)
Query: 56 PPLVQQRPQSPYQPMCPQKPRQQAPRQSI---PQNQVPQKFI-PQNQVQKASQCDPIPMK 111
P QQ+ Q P QP+ PQ P+ Q P+Q + PQ Q PQ+ + PQ Q + P+ M+
Sbjct: 784 PVAPQQQYQQPQQPVAPQ-PQYQQPQQPVAPQPQYQQPQQPVAPQPQ---DTLLHPLLMR 839
Query: 112 YADLLPILLKKNLVQTLPLPRVPNSLPP 139
D P+ T PLP + PP
Sbjct: 840 NGDSRPL-----HKPTTPLPSLDLLTPP 862
Score = 32.0 bits (71), Expect = 2.4
Identities = 32/114 (28%), Positives = 42/114 (36%), Gaps = 16/114 (14%)
Query: 189 QQQHLAPHSVAAVRPI-----TNVVQDPGYQPQFRP--SQQQYPDSHFVTTILPIMNVVR 241
QQQ+ P A +P V P YQ +P QQQY P +
Sbjct: 749 QQQYQQPQQPVAPQPQYQQPQQQVAPQPQYQQPQQPVAPQQQYQQPQQPVAPQPQYQQPQ 808
Query: 242 NP-GYQPQFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIP 294
P QPQ+QQ QQ Q P+ M+ G+ P +P P+P
Sbjct: 809 QPVAPQPQYQQPQQPVAPQPQDTLLHPLLMRNGDSRPL--------HKPTTPLP 854
>ZIPA_ECOL6 (Q8FFC0) Cell division protein zipA
Length = 332
Score = 38.9 bits (89), Expect = 0.019
Identities = 25/77 (32%), Positives = 34/77 (43%), Gaps = 8/77 (10%)
Query: 200 AVRPITNVVQDPGY------QPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQ 253
A RP P Y QP +P + Q P H P+ V+ P YQPQ +Q
Sbjct: 83 AARPSPQHQYQPPYASAQPRQPVQQPPEAQVPPQHAPRPTQPVQQPVQQPAYQPQPEQPL 142
Query: 254 QQPRQQAPRIKFDPIPM 270
QQP +P++ P P+
Sbjct: 143 QQP--VSPQVASAPQPV 157
Score = 31.6 bits (70), Expect = 3.1
Identities = 16/53 (30%), Positives = 22/53 (41%)
Query: 57 PLVQQRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIP 109
P Q + Q PY P++P QQ P +P P+ P Q + P P
Sbjct: 86 PSPQHQYQPPYASAQPRQPVQQPPEAQVPPQHAPRPTQPVQQPVQQPAYQPQP 138
>HOG3_HORVU (P80198) Gamma-hordein 3
Length = 289
Score = 38.5 bits (88), Expect = 0.025
Identities = 23/60 (38%), Positives = 30/60 (49%), Gaps = 11/60 (18%)
Query: 56 PPLVQQRPQSPY---QPMCPQKPRQQAPR--------QSIPQNQVPQKFIPQNQVQKASQ 104
PP +QQ P+ PY QP+ Q+P Q P+ Q +PQ Q PQ+ Q Q Q Q
Sbjct: 32 PPFLQQEPEQPYPQQQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPLQPQQQFPQQ 91
Score = 30.8 bits (68), Expect = 5.3
Identities = 25/81 (30%), Positives = 33/81 (39%), Gaps = 1/81 (1%)
Query: 209 QDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPRIKFDPI 268
Q P Q Q P Q Q P H LP + QPQ Q QQ P Q + +F P
Sbjct: 46 QQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPLQPQQQFPQQMPLQPQQQPQF-PQ 104
Query: 269 PMKYGELFPYLLERNLVQTRP 289
+G+ L ++ Q +P
Sbjct: 105 QKPFGQYQQPLTQQPYPQQQP 125
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,421,232
Number of Sequences: 164201
Number of extensions: 2054288
Number of successful extensions: 10493
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 123
Number of HSP's that attempted gapping in prelim test: 8993
Number of HSP's gapped (non-prelim): 885
length of query: 348
length of database: 59,974,054
effective HSP length: 111
effective length of query: 237
effective length of database: 41,747,743
effective search space: 9894215091
effective search space used: 9894215091
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146784.3


