

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.7 + phase: 0
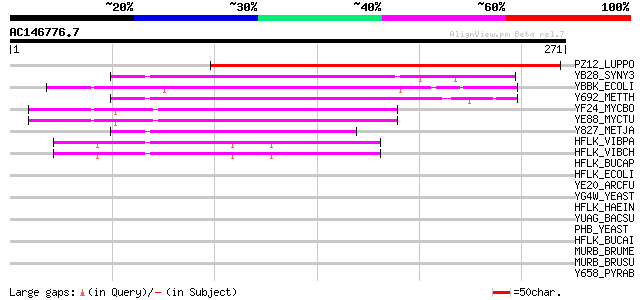
(271 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PZ12_LUPPO (P16148) PPLZ12 protein 218 1e-56
YB28_SYNY3 (P72655) Hypothetical protein slr1128 66 1e-10
YBBK_ECOLI (P77367) Hypothetical protein ybbK 63 9e-10
Y692_METTH (O26788) Hypothetical protein MTH692 55 1e-07
YF24_MYCBO (P63694) Hypothetical protein Mb1524 49 2e-05
YE88_MYCTU (P63693) Hypothetical protein Rv1488/MT1533.2 49 2e-05
Y827_METJA (Q58237) Hypothetical protein MJ0827 47 4e-05
HFLK_VIBPA (P40605) HflK protein 45 2e-04
HFLK_VIBCH (Q9KV09) HflK protein 43 7e-04
HFLK_BUCAP (Q8K914) HflK protein 40 0.005
HFLK_ECOLI (P25662) HflK protein 39 0.018
YE20_ARCFU (O28852) Hypothetical protein AF1420 37 0.067
YG4W_YEAST (P50085) Hypothetical 34.9 kDa protein in SMI1-PHO81 ... 36 0.088
HFLK_HAEIN (P44546) HflK protein 36 0.088
YUAG_BACSU (O32076) Hypothetical protein yuaG 36 0.11
PHB_YEAST (P40961) Prohibitin 36 0.11
HFLK_BUCAI (P57631) HflK protein 36 0.11
MURB_BRUME (Q8YI64) UDP-N-acetylenolpyruvoylglucosamine reductas... 35 0.15
MURB_BRUSU (Q8FZP4) UDP-N-acetylenolpyruvoylglucosamine reductas... 35 0.20
Y658_PYRAB (Q9V0Y1) Hypothetical protein PYRAB06580 34 0.33
>PZ12_LUPPO (P16148) PPLZ12 protein
Length = 184
Score = 218 bits (556), Expect = 1e-56
Identities = 104/171 (60%), Positives = 136/171 (78%)
Query: 99 LNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMR 158
+NLDD FEQK E+AK+V EE EK M YGY I L+ DI PD V+ AMNEINAA RM+
Sbjct: 1 MNLDDLFEQKGEVAKSVLEELEKVMGEYGYNIEHILMVDIIPDDSVRRAMNEINAAQRMQ 60
Query: 159 IAANEKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAK 218
+A+ K EAEKILQ+KRAE EAE+KYL G+G+ARQRQAI DGLR++++ FS V G SAK
Sbjct: 61 LASLYKGEAEKILQVKRAEAEAEAKYLGGVGVARQRQAITDGLRENILNFSHKVEGTSAK 120
Query: 219 DVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
+VMD++++TQYFDT+K++G +SK++ VFIPHGPG VRD+ QI +GL++ +
Sbjct: 121 EVMDLIMITQYFDTIKDLGNSSKNTTVFIPHGPGHVRDIGEQIRNGLMESA 171
>YB28_SYNY3 (P72655) Hypothetical protein slr1128
Length = 321
Score = 65.9 bits (159), Expect = 1e-10
Identities = 49/212 (23%), Positives = 101/212 (47%), Gaps = 18/212 (8%)
Query: 50 RDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKN 109
+DNV + A + +R + A+YK+ N + + V IR+ + KL LD TF +
Sbjct: 76 KDNVAITADAVVYWRII--DMEKAYYKVENLQSAMVNLVLTQIRSEIGKLELDQTFTART 133
Query: 110 EIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEK 169
EI + + E + + +G ++ + + DI P + V +M A R + AA +E ++
Sbjct: 134 EINELLLRELDISTDPWGVKVTRVELRDIMPSKAVLDSMELQMTAERKKRAAILTSEGQR 193
Query: 170 ILQIKRAEGEAESKYLSGMGIARQRQAIVD-------------GLRDSVIGFSENVPGPS 216
I A+G+A+++ L A+++ AI++ +++ +E + +
Sbjct: 194 DSAINSAQGDAQARVLEAE--AKKKAAILNAEAEQQKKVLEAKATAEALSILTEKLSSDN 251
Query: 217 -AKDVMDMVLVTQYFDTMKEIGAASKSSAVFI 247
A++ + +L QY + IG++ S +F+
Sbjct: 252 HAREALQFLLAQQYLNMGTTIGSSDSSKVMFL 283
>YBBK_ECOLI (P77367) Hypothetical protein ybbK
Length = 305
Score = 62.8 bits (151), Expect = 9e-10
Identities = 59/251 (23%), Positives = 111/251 (43%), Gaps = 25/251 (9%)
Query: 19 EGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLRDNVFVNVVASIQYRALANKANDAF---- 74
E FG++ K L PG + F+ RI +++ + V + + AN DA
Sbjct: 32 ERFGRYTKTLQPGLSLVVPFMD-RIGRKINMMEQVLDIPSQEVISKDNANVTIDAVCFIQ 90
Query: 75 --------YKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAY 126
Y++SN I IR + + LD+ Q++ I + ++A + +
Sbjct: 91 VIDAPRAAYEVSNLELAIINLTMTNIRTVLGSMELDEMLSQRDSINSRLLRIVDEATNPW 150
Query: 127 GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAESKYLS 186
G ++ + I D+ P + ++MN A R + A +AE + +I +AEGE +S+ L
Sbjct: 151 GIKVTRIEIRDVRPPAELISSMNAQMKAERTKRAYILEAEGIRQAEILKAEGEKQSQILK 210
Query: 187 GMG---------IARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVTQYFDTMKEIG 237
G AR+R A + ++ SE + + V + + +Y + +++IG
Sbjct: 211 AEGERQSAFLQAEARERSAEAEARATKMV--SEAIASGDIQAV-NYFVAQKYTEALQQIG 267
Query: 238 AASKSSAVFIP 248
++S S V +P
Sbjct: 268 SSSNSKVVMMP 278
>Y692_METTH (O26788) Hypothetical protein MTH692
Length = 318
Score = 55.5 bits (132), Expect = 1e-07
Identities = 50/211 (23%), Positives = 93/211 (43%), Gaps = 19/211 (9%)
Query: 50 RDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKN 109
+DN V V I Y + +A Y + + I +R + L LD T +
Sbjct: 74 KDNTVVVVDCVIFYEVV--DPFNAVYNVVDFYQAITKLAQTNLRNIIGDLELDQTLTSRE 131
Query: 110 EIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEK 169
I + E ++A +G +V+ I IEP + AM++ A RM+ AA +AE K
Sbjct: 132 MINTQLREVLDEATDKWGTRVVRVEIQRIEPPGDIVEAMSKQMKAERMKRAAILEAEGYK 191
Query: 170 ILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDM------ 223
+IKRAEG+ ++ L G A + + D + I +E AK ++ +
Sbjct: 192 QSEIKRAEGDKQAAILEAEGKAEAIKKVADANKYREIAIAEG----QAKAILSVFRAMHE 247
Query: 224 ------VLVTQYFDTMKEIGAASKSSAVFIP 248
++ +Y + ++++ A +++ + +P
Sbjct: 248 GDPTNDIIALKYLEALEKV-ADGRATKILLP 277
>YF24_MYCBO (P63694) Hypothetical protein Mb1524
Length = 381
Score = 48.5 bits (114), Expect = 2e-05
Identities = 38/194 (19%), Positives = 87/194 (44%), Gaps = 17/194 (8%)
Query: 10 VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR--------------DNVFV 55
+ Q++ A+ E G++ + + + F+ R+ + LR DN+ +
Sbjct: 29 IPQAEAAVIERLGRYSRTVSGQLTLLVPFID-RVRARVDLRERVVSFPPQPVITEDNLTL 87
Query: 56 NVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 115
N+ + ++ +A A Y++SN ++ +R V + L+ T +++I +
Sbjct: 88 NIDTVVYFQVTVPQA--AVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQL 145
Query: 116 EEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKR 175
++A +G + + + I+P ++ +M + A R + A AE + IK+
Sbjct: 146 RGVLDEATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTREAAIKQ 205
Query: 176 AEGEAESKYLSGMG 189
AEG+ +++ L+ G
Sbjct: 206 AEGQKQAQILAAEG 219
>YE88_MYCTU (P63693) Hypothetical protein Rv1488/MT1533.2
Length = 381
Score = 48.5 bits (114), Expect = 2e-05
Identities = 38/194 (19%), Positives = 87/194 (44%), Gaps = 17/194 (8%)
Query: 10 VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR--------------DNVFV 55
+ Q++ A+ E G++ + + + F+ R+ + LR DN+ +
Sbjct: 29 IPQAEAAVIERLGRYSRTVSGQLTLLVPFID-RVRARVDLRERVVSFPPQPVITEDNLTL 87
Query: 56 NVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 115
N+ + ++ +A A Y++SN ++ +R V + L+ T +++I +
Sbjct: 88 NIDTVVYFQVTVPQA--AVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQL 145
Query: 116 EEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKR 175
++A +G + + + I+P ++ +M + A R + A AE + IK+
Sbjct: 146 RGVLDEATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTREAAIKQ 205
Query: 176 AEGEAESKYLSGMG 189
AEG+ +++ L+ G
Sbjct: 206 AEGQKQAQILAAEG 219
>Y827_METJA (Q58237) Hypothetical protein MJ0827
Length = 199
Score = 47.4 bits (111), Expect = 4e-05
Identities = 31/120 (25%), Positives = 57/120 (46%), Gaps = 2/120 (1%)
Query: 50 RDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKN 109
+DN V V A + YR + A ++ + I +RA + + LD+ ++
Sbjct: 80 KDNAVVKVDAVVYYRVI--DVEKAILEVEDYEYAIINLAQTTLRAIIGSMELDEVLNKRE 137
Query: 110 EIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEK 169
I + E ++ A+G I + + +I+P + +K AM + A R++ AA +AE EK
Sbjct: 138 YINSKLLEILDRETDAWGVRIEKVEVKEIDPPEDIKNAMAQQMKAERLKRAAILEAEGEK 197
>HFLK_VIBPA (P40605) HflK protein
Length = 400
Score = 44.7 bits (104), Expect = 2e-04
Identities = 39/175 (22%), Positives = 73/175 (41%), Gaps = 17/175 (9%)
Query: 22 GKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNVVASIQYRALANK 69
GK+++++ PG + P F+ + R +G + +D V V +QYR
Sbjct: 105 GKYDRIVDPGLNWRPRFIDEYEAVNVQAIRSLRASGLMLTKDENVVTVAMDVQYRVA--D 162
Query: 70 ANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ-KNEIAKAVEEEREKAMSAY-- 126
Y+++N ++ +RA + +D + +I ++ +E + + +Y
Sbjct: 163 PYKYLYRVTNADDSLRQATDSALRAVIGDSLMDSILTSGRQQIRQSTQETLNQIIDSYDM 222
Query: 127 GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAE 181
G IV P + VK A ++ AA +AEA K + +A G AE
Sbjct: 223 GLVIVDVNFQSARPPEQVKDAFDDAIAAREDEERFIREAEAYKNEILPKATGRAE 277
>HFLK_VIBCH (Q9KV09) HflK protein
Length = 395
Score = 43.1 bits (100), Expect = 7e-04
Identities = 39/175 (22%), Positives = 73/175 (41%), Gaps = 17/175 (9%)
Query: 22 GKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNVVASIQYRALANK 69
GK+++++ PG + P F+ + R +G + +D V V +QYR
Sbjct: 102 GKYDRIVDPGLNWRPRFIDEVTPVNVQAIRSLRASGLMLTKDENVVTVSMDVQYRIA--D 159
Query: 70 ANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ-KNEIAKAVEEEREKAMSAY-- 126
Y+++N ++ +RA V +D + +I ++ ++ + + +Y
Sbjct: 160 PYKYLYRVTNADDSLRQATDSALRAVVGDSLMDSILTSGRQQIRQSTQQTLNQVIDSYDM 219
Query: 127 GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAE 181
G IV P + VK A ++ AA +AEA K + +A G AE
Sbjct: 220 GLMIVDVNFQSARPPEQVKDAFDDAIAAREDEERFIREAEAYKNEILPKATGRAE 274
>HFLK_BUCAP (Q8K914) HflK protein
Length = 411
Score = 40.4 bits (93), Expect = 0.005
Identities = 51/242 (21%), Positives = 96/242 (39%), Gaps = 24/242 (9%)
Query: 21 FGKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNVVASIQYRALAN 68
FGKF ++ PG + P F+ + +G + D V V ++QY+
Sbjct: 102 FGKFSHLVAPGLNWRPVFINEVKAVNVETVRELATSGVMLTSDENVVRVEMNVQYK--IT 159
Query: 69 KANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTF-EQKNEIAKAVEEEREKAMSAY- 126
D + ++ ++ +R + N+D E + I ++E E+ + Y
Sbjct: 160 DPADYLFSVAYPDDSLRQATDSALRGVIGHSNMDRVLTEGRTLIRSDTQKEIEETIKPYK 219
Query: 127 -GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAESKYL 185
G I+ P + VK A ++ AA R +AEA +A G+A+
Sbjct: 220 LGITILDVNFQTARPPEEVKEAFDDAIAARENREQYIREAEAYSNEVQPKAHGKAQRILE 279
Query: 186 SGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVTQYFDTMKEIGAASKSSAV 245
+ +R G V+ F + + P + +M L Y ++M+++ SK+ +
Sbjct: 280 EAKAYSSRRILEAQG---EVVRFLKIL--PEYRKNKEMTLKRLYIESMEKL--LSKTKKI 332
Query: 246 FI 247
FI
Sbjct: 333 FI 334
>HFLK_ECOLI (P25662) HflK protein
Length = 419
Score = 38.5 bits (88), Expect = 0.018
Identities = 48/231 (20%), Positives = 91/231 (38%), Gaps = 22/231 (9%)
Query: 21 FGKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNVVASIQYRALAN 68
FGKF ++ PG + P F+ + +G + D V V ++QYR
Sbjct: 111 FGKFSHLVEPGLNWKPTFIDEVKPVNVEAVRELAASGVMLTSDENVVRVEMNVQYR--VT 168
Query: 69 KANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTF-EQKNEIAKAVEEEREKAMSAY- 126
Y +++ ++ +R + K +D E + I + E E+ + Y
Sbjct: 169 NPEKYLYSVTSPDDSLRQATDSALRGVIGKYTMDRILTEGRTVIRSDTQRELEETIRPYD 228
Query: 127 -GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAESKYL 185
G ++ P + VK A ++ AA +AEA RA G+A+ +
Sbjct: 229 MGITLLDVNFQAARPPEEVKAAFDDAIAARENEQQYIREAEAYTNEVQPRANGQAQR--I 286
Query: 186 SGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVTQYFDTMKEI 236
A + Q I++ + V F++ + P K ++ Y +TM+++
Sbjct: 287 LEEARAYKAQTILEA-QGEVARFAKLL--PEYKAAPEITRERLYIETMEKV 334
>YE20_ARCFU (O28852) Hypothetical protein AF1420
Length = 249
Score = 36.6 bits (83), Expect = 0.067
Identities = 30/135 (22%), Positives = 62/135 (45%), Gaps = 17/135 (12%)
Query: 50 RDNVFVNVVASIQYRAL--ANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
+DNV V V A + YR + A + F T Q +R+ + + LD+ +
Sbjct: 75 KDNVTVKVNAVVYYRVVDPAKAVTEVFDYQYATAQLAQT----TLRSIIGQAELDEVLSE 130
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
++++ +++ ++ + +G ++ I D+E + ++ M A +AE
Sbjct: 131 RDKLNVKLQQIIDEETNPWGIKVTAVEIKDVELPEEMRRIM-----------AMQAEAER 179
Query: 168 EKILQIKRAEGEAES 182
E+ +I RAEGE ++
Sbjct: 180 ERRSKIIRAEGEYQA 194
>YG4W_YEAST (P50085) Hypothetical 34.9 kDa protein in SMI1-PHO81
intergenic region
Length = 315
Score = 36.2 bits (82), Expect = 0.088
Identities = 27/117 (23%), Positives = 51/117 (43%), Gaps = 3/117 (2%)
Query: 84 IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQH 143
+ + V +V++A V + N Q+ ++++ + E + S + + IT +
Sbjct: 150 LPSIVNEVLKAVVAQFNASQLITQREKVSRLIRENLVRRASKFNILLDDVSITYMTFSPE 209
Query: 144 VKTAMNEINAA---ARMRIAANEKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAI 197
A+ A A+ +KA EK + RA+GEA+S L G I + R +
Sbjct: 210 FTNAVEAKQIAQQDAQRAAFVVDKARQEKQGMVVRAQGEAKSAELIGEAIKKSRDYV 266
>HFLK_HAEIN (P44546) HflK protein
Length = 410
Score = 36.2 bits (82), Expect = 0.088
Identities = 47/242 (19%), Positives = 89/242 (36%), Gaps = 40/242 (16%)
Query: 21 FGKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNVVASIQYRALAN 68
FG+ ++ PG + P F+ K R G + +D V V ++QYR
Sbjct: 118 FGELHSIVQPGLNWKPTFVDKVLPVNVEQVKELRTQGAMLTQDENMVKVEMTVQYR--VQ 175
Query: 69 KANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSA--- 125
+ ++N + +R + ++++D + V E KA++
Sbjct: 176 DPAKYLFSVTNADDSLNQATDSALRYVIGHMSMNDILTTGRSV---VRENTWKALNEIIK 232
Query: 126 ---YGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAES 182
G E++ P + VK A ++ KA+ ++ I+ AE A
Sbjct: 233 SYDMGLEVIDVNFQSARPPEEVKDAFDDA-----------IKAQEDEQRFIREAEAYARE 281
Query: 183 KYLSGMGIARQRQAIVDGLRDSVI----GFSENVPG--PSAKDVMDMVLVTQYFDTMKEI 236
K G A++ +D ++ G E + P K D++ Y TM+++
Sbjct: 282 KEPIARGDAQRILEEATAYKDRIVLDAKGEVERLQRLLPEFKAAPDLLRERLYIQTMEKV 341
Query: 237 GA 238
A
Sbjct: 342 MA 343
>YUAG_BACSU (O32076) Hypothetical protein yuaG
Length = 509
Score = 35.8 bits (81), Expect = 0.11
Identities = 20/92 (21%), Positives = 46/92 (49%)
Query: 92 IRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEI 151
+R+ + + +++ ++ + + ++ V+ + ++ G IV I D+ ++ +
Sbjct: 143 LRSILGSMTVEEIYKNREKFSQEVQRVASQDLAKMGLVIVSFTIKDVRDKNGYLESLGKP 202
Query: 152 NAAARMRIAANEKAEAEKILQIKRAEGEAESK 183
A R A AEA+K +IKRAE + ++K
Sbjct: 203 RIAQVKRDADIATAEADKETRIKRAEADKDAK 234
>PHB_YEAST (P40961) Prohibitin
Length = 287
Score = 35.8 bits (81), Expect = 0.11
Identities = 28/115 (24%), Positives = 53/115 (45%), Gaps = 7/115 (6%)
Query: 90 DVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN 149
+V+++ V + + + Q+ I++ + +E + +G ++ IT + A+
Sbjct: 127 EVLKSIVAQFDAAELITQREIISQKIRKELSTRANEFGIKLEDVSITHMTFGPEFTKAVE 186
Query: 150 EINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGL 201
+ A + A EKAE E+ + RAEGEAES +A+ + DGL
Sbjct: 187 QKQIAQQDAERAKFLVEKAEQERQASVIRAEGEAESAEFISKALAK----VGDGL 237
>HFLK_BUCAI (P57631) HflK protein
Length = 406
Score = 35.8 bits (81), Expect = 0.11
Identities = 37/187 (19%), Positives = 74/187 (38%), Gaps = 17/187 (9%)
Query: 10 VDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGK------------RIAGHLSLRDNVFVNV 57
+ +++ + FGKF ++ PG + P F + +G + D V V
Sbjct: 87 ITEAERGVVTSFGKFSHLVQPGLNWRPVFFNEVKPVNVETVRELATSGIMLTADENVVRV 146
Query: 58 VASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKA-VE 116
++QY+ + N A D + + ++ +R + +D + + ++ +
Sbjct: 147 EMNVQYK-ITNPA-DYLFSVCYPDDSLRQATDSALRGVIGHSTMDRVLTEGRTLVRSDTQ 204
Query: 117 EEREKAMSAY--GYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIK 174
+E E + Y G I+ P + VK A ++ AA R +AEA
Sbjct: 205 KEIENTIKPYKMGITILDVNFQTARPPEEVKAAFDDAIAARENREQYVREAEAYSNEVKP 264
Query: 175 RAEGEAE 181
+A G+A+
Sbjct: 265 KANGKAQ 271
>MURB_BRUME (Q8YI64) UDP-N-acetylenolpyruvoylglucosamine reductase
(EC 1.1.1.158) (UDP-N-acetylmuramate dehydrogenase)
Length = 322
Score = 35.4 bits (80), Expect = 0.15
Identities = 34/124 (27%), Positives = 53/124 (42%), Gaps = 5/124 (4%)
Query: 151 INAAARMRIAANEKAEAEKILQIKRAEGEAESKYLSG--MGIA-RQRQAIVDGLRDSVIG 207
I A RM AN E++++++ + + E LS MG A R A D + SV+
Sbjct: 138 IGGALRMNAGANGVETRERVVEVRALDRKGEVHVLSNADMGYAYRHSSASPDLIFTSVL- 196
Query: 208 FSENVPGPSAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQ 267
E VPG M V + +T++ + + S P G A +++ C GL
Sbjct: 197 -FEGVPGERDDIRRAMDEVQHHRETVQPVREKTGGSTFKNPEGTSAWKEIDKAGCRGLRV 255
Query: 268 GSHQ 271
G Q
Sbjct: 256 GGAQ 259
>MURB_BRUSU (Q8FZP4) UDP-N-acetylenolpyruvoylglucosamine reductase
(EC 1.1.1.158) (UDP-N-acetylmuramate dehydrogenase)
Length = 322
Score = 35.0 bits (79), Expect = 0.20
Identities = 33/124 (26%), Positives = 53/124 (42%), Gaps = 5/124 (4%)
Query: 151 INAAARMRIAANEKAEAEKILQIKRAEGEAESKYLSG--MGIA-RQRQAIVDGLRDSVIG 207
+ A RM AN E++++++ + + E LS MG A R A D + SV+
Sbjct: 138 VGGALRMNAGANGVETRERVVEVRALDRKGEVHVLSNADMGYAYRHSSASPDLIFTSVL- 196
Query: 208 FSENVPGPSAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQ 267
E VPG M V + +T++ + + S P G A +++ C GL
Sbjct: 197 -FEGVPGERDDIRRAMDEVQHHRETVQPVREKTGGSTFKNPEGTSAWKEIDKAGCRGLRV 255
Query: 268 GSHQ 271
G Q
Sbjct: 256 GGAQ 259
>Y658_PYRAB (Q9V0Y1) Hypothetical protein PYRAB06580
Length = 268
Score = 34.3 bits (77), Expect = 0.33
Identities = 28/123 (22%), Positives = 58/123 (46%), Gaps = 8/123 (6%)
Query: 50 RDNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQA---YVFDVIRASVPKLNLDDTFE 106
+DNV V V A + +R + D ++ + I A +R+ + + +LD+
Sbjct: 80 KDNVPVRVNAVVYFRVV-----DPVKAVTQVKNYIMATSQISQTTLRSVIGQAHLDELLS 134
Query: 107 QKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAE 166
+++++ ++ ++A +G ++ I D+E ++ AM + A R R A AE
Sbjct: 135 ERDKLNMQLQRIIDEATDPWGIKVTAVEIKDVELPAGMQRAMAKQAEAERERRARITLAE 194
Query: 167 AEK 169
AE+
Sbjct: 195 AER 197
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,667,885
Number of Sequences: 164201
Number of extensions: 1084898
Number of successful extensions: 3694
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 3664
Number of HSP's gapped (non-prelim): 70
length of query: 271
length of database: 59,974,054
effective HSP length: 108
effective length of query: 163
effective length of database: 42,240,346
effective search space: 6885176398
effective search space used: 6885176398
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146776.7


