

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.3 - phase: 0
(286 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
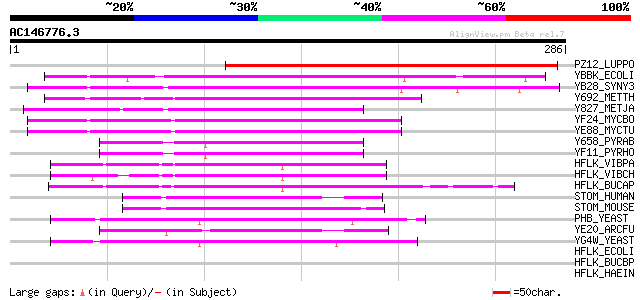
Score E
Sequences producing significant alignments: (bits) Value
PZ12_LUPPO (P16148) PPLZ12 protein 226 5e-59
YBBK_ECOLI (P77367) Hypothetical protein ybbK 78 2e-14
YB28_SYNY3 (P72655) Hypothetical protein slr1128 77 6e-14
Y692_METTH (O26788) Hypothetical protein MTH692 66 9e-11
Y827_METJA (Q58237) Hypothetical protein MJ0827 57 7e-08
YF24_MYCBO (P63694) Hypothetical protein Mb1524 55 2e-07
YE88_MYCTU (P63693) Hypothetical protein Rv1488/MT1533.2 55 2e-07
Y658_PYRAB (Q9V0Y1) Hypothetical protein PYRAB06580 52 2e-06
YF11_PYRHO (O59180) Hypothetical protein PH1511 50 5e-06
HFLK_VIBPA (P40605) HflK protein 49 1e-05
HFLK_VIBCH (Q9KV09) HflK protein 48 2e-05
HFLK_BUCAP (Q8K914) HflK protein 47 5e-05
STOM_HUMAN (P27105) Erythrocyte band 7 integral membrane protein... 46 1e-04
STOM_MOUSE (P54116) Erythrocyte band 7 integral membrane protein... 45 2e-04
PHB_YEAST (P40961) Prohibitin 45 2e-04
YE20_ARCFU (O28852) Hypothetical protein AF1420 45 3e-04
YG4W_YEAST (P50085) Hypothetical 34.9 kDa protein in SMI1-PHO81 ... 44 5e-04
HFLK_ECOLI (P25662) HflK protein 43 0.001
HFLK_BUCBP (Q89A39) HflK protein 41 0.004
HFLK_HAEIN (P44546) HflK protein 40 0.005
>PZ12_LUPPO (P16148) PPLZ12 protein
Length = 184
Score = 226 bits (576), Expect = 5e-59
Identities = 108/171 (63%), Positives = 140/171 (81%)
Query: 112 LNLDDTFEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMR 171
+NLDD FEQK E+AK+V EELEK M YGY I L+ DI PD V+RAMNEINAA RM+
Sbjct: 1 MNLDDLFEQKGEVAKSVLEELEKVMGEYGYNIEHILMVDIIPDDSVRRAMNEINAAQRMQ 60
Query: 172 LAAKEKAEAEKILQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVIGFSVNVPGTTAK 231
LA+ K EAEKILQ+KRAE EAE+KYL G+G+ARQRQAI DGLR++++ FS V GT+AK
Sbjct: 61 LASLYKGEAEKILQVKRAEAEAEAKYLGGVGVARQRQAITDGLRENILNFSHKVEGTSAK 120
Query: 232 DVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQIRDGLLQGS 282
+VMD++++TQYFDT+K++G +SK++ VFIPHGPG VRD+ QIR+GL++ +
Sbjct: 121 EVMDLIMITQYFDTIKDLGNSSKNTTVFIPHGPGHVRDIGEQIRNGLMESA 171
>YBBK_ECOLI (P77367) Hypothetical protein ybbK
Length = 305
Score = 78.2 bits (191), Expect = 2e-14
Identities = 68/274 (24%), Positives = 127/274 (45%), Gaps = 24/274 (8%)
Query: 19 EGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCE---TKTKDNVFVNVVASIQ 75
E FG++ K LQPG + F+ RI +++ Q LDI + +K NV ++ V IQ
Sbjct: 32 ERFGRYTKTLQPGLSLVVPFMD-RIGRKINMMEQVLDIPSQEVISKDNANVTIDAVCFIQ 90
Query: 76 YRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEELEKA 135
A A Y++SN I IR + + LD+ Q++ I + +++A
Sbjct: 91 ----VIDAPRAAYEVSNLELAIINLTMTNIRTVLGSMELDEMLSQRDSINSRLLRIVDEA 146
Query: 136 MSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAES 195
+ +G ++ + I D+ P + +MN A R + A +AE + +I +AEGE +S
Sbjct: 147 TNPWGIKVTRIEIRDVRPPAELISSMNAQMKAERTKRAYILEAEGIRQAEILKAEGEKQS 206
Query: 196 KYLSGLG---------IARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVTQYFDTM 246
+ L G AR+R A + ++ ++ A ++ + +Y + +
Sbjct: 207 QILKAEGERQSAFLQAEARERSAEAEARATKMVSEAIASGDIQA---VNYFVAQKYTEAL 263
Query: 247 KEIGAASKSSAVFIPHGP----GAVRDVASQIRD 276
++IG++S S V +P G++ +A ++D
Sbjct: 264 QQIGSSSNSKVVMMPLEASSLMGSIAGIAELVKD 297
>YB28_SYNY3 (P72655) Hypothetical protein slr1128
Length = 321
Score = 76.6 bits (187), Expect = 6e-14
Identities = 70/289 (24%), Positives = 130/289 (44%), Gaps = 18/289 (6%)
Query: 10 VDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETK-TKDNVFV 68
V++ + E G + K L PG + L R+ + R + +DI ++ TKDNV +
Sbjct: 23 VNEKNEYLVERLGSYNKKLTPGLNFTVPILD-RVVFKQTTREKVIDIPPQSCITKDNVAI 81
Query: 69 NVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 128
A + +R + A+YK+ N ++ + V IR+ + KL LD TF + EI + +
Sbjct: 82 TADAVVYWRII--DMEKAYYKVENLQSAMVNLVLTQIRSEIGKLELDQTFTARTEINELL 139
Query: 129 EEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKR 188
EL+ + +G ++ + + DI P V +M A R + AA +E ++ I
Sbjct: 140 LRELDISTDPWGVKVTRVELRDIMPSKAVLDSMELQMTAERKKRAAILTSEGQRDSAINS 199
Query: 189 AEGEAESKYLSG--------LGIARQRQAIVDGLRDSVIGFSVNVPGTT----AKDVMDM 236
A+G+A+++ L L ++Q V + + S+ + A++ +
Sbjct: 200 AQGDAQARVLEAEAKKKAAILNAEAEQQKKVLEAKATAEALSILTEKLSSDNHAREALQF 259
Query: 237 VLVTQYFDTMKEIGAASKSSAVFIP--HGPGAVRDVASQIRDGLLQGSL 283
+L QY + IG++ S +F+ + + V S + DG L L
Sbjct: 260 LLAQQYLNMGTTIGSSDSSKVMFLDPRNILSTLEGVRSIVGDGALDEGL 308
>Y692_METTH (O26788) Hypothetical protein MTH692
Length = 318
Score = 66.2 bits (160), Expect = 9e-11
Identities = 56/195 (28%), Positives = 92/195 (46%), Gaps = 5/195 (2%)
Query: 19 EGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKC-ETKTKDNVFVNVVASIQYR 77
E GK+++ ++ G + F+ + +R Q +D+ E TKDN V VV + +
Sbjct: 31 ERLGKYQRTVESGLVVIIPFI--EAIKKVDMREQVVDVPPQEVITKDNTVV-VVDCVIFY 87
Query: 78 ALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMS 137
+ D N A Y + + I +R + L LD T + I + E L++A
Sbjct: 88 EVVDPFN-AVYNVVDFYQAITKLAQTNLRNIIGDLELDQTLTSREMINTQLREVLDEATD 146
Query: 138 AYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAESKY 197
+G +V+ I IEP + AM++ A RM+ AA +AE K +IKRAEG+ ++
Sbjct: 147 KWGTRVVRVEIQRIEPPGDIVEAMSKQMKAERMKRAAILEAEGYKQSEIKRAEGDKQAAI 206
Query: 198 LSGLGIARQRQAIVD 212
L G A + + D
Sbjct: 207 LEAEGKAEAIKKVAD 221
>Y827_METJA (Q58237) Hypothetical protein MJ0827
Length = 199
Score = 56.6 bits (135), Expect = 7e-08
Identities = 46/175 (26%), Positives = 81/175 (46%), Gaps = 3/175 (1%)
Query: 8 VQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVF 67
V V+Q + + G+ L+PG + + FL + + RV + + E TKDN
Sbjct: 26 VIVNQYEGGLIFRLGRVIGKLKPGINIIIPFLDVPVKVDMRTRVTDIPPQ-EMITKDNAV 84
Query: 68 VNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKA 127
V V A + YR + A ++ + I +RA + + LD+ ++ I
Sbjct: 85 VKVDAVVYYRVI--DVEKAILEVEDYEYAIINLAQTTLRAIIGSMELDEVLNKREYINSK 142
Query: 128 VEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEK 182
+ E L++ A+G I + + +I+P +K AM + A R++ AA +AE EK
Sbjct: 143 LLEILDRETDAWGVRIEKVEVKEIDPPEDIKNAMAQQMKAERLKRAAILEAEGEK 197
>YF24_MYCBO (P63694) Hypothetical protein Mb1524
Length = 381
Score = 55.5 bits (132), Expect = 2e-07
Identities = 40/194 (20%), Positives = 93/194 (47%), Gaps = 4/194 (2%)
Query: 10 VDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETK-TKDNVFV 68
+ Q++ A+ E G++ + + + F+ R+ + LR + + + T+DN+ +
Sbjct: 29 IPQAEAAVIERLGRYSRTVSGQLTLLVPFID-RVRARVDLRERVVSFPPQPVITEDNLTL 87
Query: 69 NVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 128
N+ + ++ +A A Y++SN ++ +R V + L+ T +++I +
Sbjct: 88 NIDTVVYFQVTVPQA--AVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQL 145
Query: 129 EEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKR 188
L++A +G + + + I+P ++ +M + A R + A AE + IK+
Sbjct: 146 RGVLDEATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTREAAIKQ 205
Query: 189 AEGEAESKYLSGLG 202
AEG+ +++ L+ G
Sbjct: 206 AEGQKQAQILAAEG 219
>YE88_MYCTU (P63693) Hypothetical protein Rv1488/MT1533.2
Length = 381
Score = 55.5 bits (132), Expect = 2e-07
Identities = 40/194 (20%), Positives = 93/194 (47%), Gaps = 4/194 (2%)
Query: 10 VDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETK-TKDNVFV 68
+ Q++ A+ E G++ + + + F+ R+ + LR + + + T+DN+ +
Sbjct: 29 IPQAEAAVIERLGRYSRTVSGQLTLLVPFID-RVRARVDLRERVVSFPPQPVITEDNLTL 87
Query: 69 NVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIAKAV 128
N+ + ++ +A A Y++SN ++ +R V + L+ T +++I +
Sbjct: 88 NIDTVVYFQVTVPQA--AVYEISNYIVGVEQLTTTTLRNVVGGMTLEQTLTSRDQINAQL 145
Query: 129 EEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKR 188
L++A +G + + + I+P ++ +M + A R + A AE + IK+
Sbjct: 146 RGVLDEATGRWGLRVARVELRSIDPPPSIQASMEKQMKADREKRAMILTAEGTREAAIKQ 205
Query: 189 AEGEAESKYLSGLG 202
AEG+ +++ L+ G
Sbjct: 206 AEGQKQAQILAAEG 219
>Y658_PYRAB (Q9V0Y1) Hypothetical protein PYRAB06580
Length = 268
Score = 52.0 bits (123), Expect = 2e-06
Identities = 39/140 (27%), Positives = 71/140 (49%), Gaps = 9/140 (6%)
Query: 47 LSLRVQQLDIKC-ETKTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQA---YVF 102
+ LR Q LD+ ET TKDNV V V A + +R + D ++ +N I A
Sbjct: 63 VDLRTQVLDVPVQETITKDNVPVRVNAVVYFRVV-----DPVKAVTQVKNYIMATSQISQ 117
Query: 103 DVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMN 162
+R+ + + +LD+ +++++ ++ +++A +G ++ I D+E ++RAM
Sbjct: 118 TTLRSVIGQAHLDELLSERDKLNMQLQRIIDEATDPWGIKVTAVEIKDVELPAGMQRAMA 177
Query: 163 EINAAARMRLAAKEKAEAEK 182
+ A R R A AEAE+
Sbjct: 178 KQAEAERERRARITLAEAER 197
>YF11_PYRHO (O59180) Hypothetical protein PH1511
Length = 266
Score = 50.4 bits (119), Expect = 5e-06
Identities = 38/140 (27%), Positives = 70/140 (49%), Gaps = 9/140 (6%)
Query: 47 LSLRVQQLDIKC-ETKTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQA---YVF 102
+ LR Q LD+ ET TKDNV V V A + +R + D ++ +N I A
Sbjct: 63 VDLRTQVLDVPVQETITKDNVPVRVNAVVYFRVV-----DPVKAVTQVKNYIMATSQISQ 117
Query: 103 DVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMN 162
+R+ + + +LD+ +++++ ++ +++A +G ++ I D+E +++AM
Sbjct: 118 TTLRSVIGQAHLDELLSERDKLNMQLQRIIDEATDPWGIKVTAVEIKDVELPAGMQKAMA 177
Query: 163 EINAAARMRLAAKEKAEAEK 182
A R R A AEAE+
Sbjct: 178 RQAEAERERRARITLAEAER 197
>HFLK_VIBPA (P40605) HflK protein
Length = 400
Score = 49.3 bits (116), Expect = 1e-05
Identities = 44/176 (25%), Positives = 80/176 (45%), Gaps = 6/176 (3%)
Query: 22 GKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALAD 81
GK+++++ PG + P F+ + A ++ ++ L TKD V V +QYR +AD
Sbjct: 105 GKYDRIVDPGLNWRPRFIDEYEAVNVQ-AIRSLRASGLMLTKDENVVTVAMDVQYR-VAD 162
Query: 82 KANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ-KNEIAKAVEEELEKAMSAY- 139
Y+++N + ++ +RA + +D + +I ++ +E L + + +Y
Sbjct: 163 PYK-YLYRVTNADDSLRQATDSALRAVIGDSLMDSILTSGRQQIRQSTQETLNQIIDSYD 221
Query: 140 -GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAE 194
G IV P VK A ++ AA +AEA K + +A G AE
Sbjct: 222 MGLVIVDVNFQSARPPEQVKDAFDDAIAAREDEERFIREAEAYKNEILPKATGRAE 277
>HFLK_VIBCH (Q9KV09) HflK protein
Length = 395
Score = 48.1 bits (113), Expect = 2e-05
Identities = 46/180 (25%), Positives = 80/180 (43%), Gaps = 14/180 (7%)
Query: 22 GKFEKVLQPGCHCMPWFLGK----RIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYR 77
GK+++++ PG + P F+ + + SLR L + TKD V V +QYR
Sbjct: 102 GKYDRIVDPGLNWRPRFIDEVTPVNVQAIRSLRASGLML-----TKDENVVTVSMDVQYR 156
Query: 78 ALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ-KNEIAKAVEEELEKAM 136
+AD Y+++N + ++ +RA V +D + +I ++ ++ L + +
Sbjct: 157 -IADPYK-YLYRVTNADDSLRQATDSALRAVVGDSLMDSILTSGRQQIRQSTQQTLNQVI 214
Query: 137 SAY--GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAE 194
+Y G IV P VK A ++ AA +AEA K + +A G AE
Sbjct: 215 DSYDMGLMIVDVNFQSARPPEQVKDAFDDAIAAREDEERFIREAEAYKNEILPKATGRAE 274
>HFLK_BUCAP (Q8K914) HflK protein
Length = 411
Score = 47.0 bits (110), Expect = 5e-05
Identities = 56/243 (23%), Positives = 104/243 (42%), Gaps = 13/243 (5%)
Query: 21 FGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALA 80
FGKF ++ PG + P F+ + A ++ V++L T D V V ++QY+ +
Sbjct: 102 FGKFSHLVAPGLNWRPVFINEVKAVNVE-TVRELATSGVMLTSDENVVRVEMNVQYK-IT 159
Query: 81 DKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTF-EQKNEIAKAVEEELEKAMSAY 139
D A D + ++ + ++ +R + N+D E + I ++E+E+ + Y
Sbjct: 160 DPA-DYLFSVAYPDDSLRQATDSALRGVIGHSNMDRVLTEGRTLIRSDTQKEIEETIKPY 218
Query: 140 --GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAESKY 197
G I+ P VK A ++ AA R +AEA +A G+A+
Sbjct: 219 KLGITILDVNFQTARPPEEVKEAFDDAIAARENREQYIREAEAYSNEVQPKAHGKAQRIL 278
Query: 198 LSGLGIARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVTQYFDTMKEIGAASKSSA 257
+ +R G V+ F +P + +M L Y ++M+++ SK+
Sbjct: 279 EEAKAYSSRRILEAQG---EVVRFLKILP--EYRKNKEMTLKRLYIESMEKL--LSKTKK 331
Query: 258 VFI 260
+FI
Sbjct: 332 IFI 334
>STOM_HUMAN (P27105) Erythrocyte band 7 integral membrane protein
(Stomatin) (Protein 7.2b)
Length = 287
Score = 45.8 bits (107), Expect = 1e-04
Identities = 35/134 (26%), Positives = 61/134 (45%), Gaps = 13/134 (9%)
Query: 59 ETKTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTF 118
E TKD+V ++V + YR A A ++N + + +R + NL
Sbjct: 106 EILTKDSVTISVDGVVYYRV--QNATLAVANITNADSATRLLAQTTLRNVLGTKNLSQIL 163
Query: 119 EQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKA 178
+ EIA ++ L+ A A+G ++ + I D++ V ++RAM AA+ +A
Sbjct: 164 SDREEIAHNMQSTLDDATDAWGIKVERVEIKDVKLPVQLQRAM-----------AAEAEA 212
Query: 179 EAEKILQIKRAEGE 192
E ++ AEGE
Sbjct: 213 SREARAKVIAAEGE 226
>STOM_MOUSE (P54116) Erythrocyte band 7 integral membrane protein
(Stomatin) (Protein 7.2b)
Length = 284
Score = 45.4 bits (106), Expect = 2e-04
Identities = 37/135 (27%), Positives = 61/135 (44%), Gaps = 4/135 (2%)
Query: 59 ETKTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTF 118
E TKD+V ++V + YR A A ++N + + +R ++ NL
Sbjct: 107 EVLTKDSVTISVDGVVYYRV--QNATLAVANITNADSATRLLAQTTLRNALGTKNLSQIL 164
Query: 119 EQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKA 178
+ EIA ++ L+ A +G ++ + I D++ V ++RAM AAR A A
Sbjct: 165 SDREEIAHHMQSTLDDATDDWGIKVERVEIKDVKLPVQLQRAMAAEAEAAREARAKVIAA 224
Query: 179 EAEKILQIKRAEGEA 193
E E + RA EA
Sbjct: 225 EGE--MNASRALKEA 237
>PHB_YEAST (P40961) Prohibitin
Length = 287
Score = 45.4 bits (106), Expect = 2e-04
Identities = 46/198 (23%), Positives = 89/198 (44%), Gaps = 11/198 (5%)
Query: 22 GKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALAD 81
G ++V+ G H + +L K I +R + I T TKD V++ + +R
Sbjct: 46 GVKQQVVGEGTHFLVPWLQKAII--YDVRTKPKSIATNTGTKDLQMVSLTLRVLHRPEVL 103
Query: 82 KANDAFYKLSNTRNQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAY 139
+ + L ++ + + +V+++ V + + + Q+ I++ + +EL + +
Sbjct: 104 QLPAIYQNLGLDYDERVLPSIGNEVLKSIVAQFDAAELITQREIISQKIRKELSTRANEF 163
Query: 140 GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAK---EKAEAEKILQIKRAEGEAESK 196
G ++ IT + +A+ + A + AK EKAE E+ + RAEGEAES
Sbjct: 164 GIKLEDVSITHMTFGPEFTKAVEQKQIAQQDAERAKFLVEKAEQERQASVIRAEGEAESA 223
Query: 197 YLSGLGIARQRQAIVDGL 214
+A+ + DGL
Sbjct: 224 EFISKALAK----VGDGL 237
>YE20_ARCFU (O28852) Hypothetical protein AF1420
Length = 249
Score = 44.7 bits (104), Expect = 3e-04
Identities = 37/152 (24%), Positives = 71/152 (46%), Gaps = 18/152 (11%)
Query: 47 LSLRVQQLDIKC-ETKTKDNVFVNVVASIQYRAL--ADKANDAFYKLSNTRNQIQAYVFD 103
+ LR D+ E TKDNV V V A + YR + A + F T Q
Sbjct: 58 VDLRTVTYDVPSQEVVTKDNVTVKVNAVVYYRVVDPAKAVTEVFDYQYATAQLAQT---- 113
Query: 104 VIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNE 163
+R+ + + LD+ +++++ +++ +++ + +G ++ I D+E ++R M
Sbjct: 114 TLRSIIGQAELDEVLSERDKLNVKLQQIIDEETNPWGIKVTAVEIKDVELPEEMRRIM-- 171
Query: 164 INAAARMRLAAKEKAEAEKILQIKRAEGEAES 195
A + +AE E+ +I RAEGE ++
Sbjct: 172 ---------AMQAEAERERRSKIIRAEGEYQA 194
>YG4W_YEAST (P50085) Hypothetical 34.9 kDa protein in SMI1-PHO81
intergenic region
Length = 315
Score = 43.9 bits (102), Expect = 5e-04
Identities = 43/195 (22%), Positives = 80/195 (40%), Gaps = 9/195 (4%)
Query: 22 GKFEKVLQPGCHCM-PWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALA 80
G ++ G H + PW I +R + ++ T TKD VN+ + R
Sbjct: 75 GVSSRIFNEGTHFIFPWLDTPII---YDVRAKPRNVASLTGTKDLQMVNITCRVLSRPDV 131
Query: 81 DKANDAFYKLSNTRNQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEELEKAMSA 138
+ + L ++ + + V +V++A V + N Q+ ++++ + E L + S
Sbjct: 132 VQLPTIYRTLGQDYDERVLPSIVNEVLKAVVAQFNASQLITQREKVSRLIRENLVRRASK 191
Query: 139 YGYEIVQTLITDIEPDVHVKRAMNEINAA---ARMRLAAKEKAEAEKILQIKRAEGEAES 195
+ + IT + A+ A A+ +KA EK + RA+GEA+S
Sbjct: 192 FNILLDDVSITYMTFSPEFTNAVEAKQIAQQDAQRAAFVVDKARQEKQGMVVRAQGEAKS 251
Query: 196 KYLSGLGIARQRQAI 210
L G I + R +
Sbjct: 252 AELIGEAIKKSRDYV 266
>HFLK_ECOLI (P25662) HflK protein
Length = 419
Score = 42.7 bits (99), Expect = 0.001
Identities = 41/177 (23%), Positives = 73/177 (41%), Gaps = 6/177 (3%)
Query: 21 FGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALA 80
FGKF +++PG + P F+ + ++ V++L T D V V ++QYR
Sbjct: 111 FGKFSHLVEPGLNWKPTFIDEVKPVNVE-AVRELAASGVMLTSDENVVRVEMNVQYR--V 167
Query: 81 DKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTF-EQKNEIAKAVEEELEKAMSAY 139
Y +++ + ++ +R + K +D E + I + ELE+ + Y
Sbjct: 168 TNPEKYLYSVTSPDDSLRQATDSALRGVIGKYTMDRILTEGRTVIRSDTQRELEETIRPY 227
Query: 140 --GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAE 194
G ++ P VK A ++ AA +AEA RA G+A+
Sbjct: 228 DMGITLLDVNFQAARPPEEVKAAFDDAIAARENEQQYIREAEAYTNEVQPRANGQAQ 284
>HFLK_BUCBP (Q89A39) HflK protein
Length = 417
Score = 40.8 bits (94), Expect = 0.004
Identities = 36/146 (24%), Positives = 68/146 (45%), Gaps = 12/146 (8%)
Query: 10 VDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVN 69
+ +S+ + FGKF + PG H P + K I +S V++++ T FV
Sbjct: 91 IQESEYGVVTCFGKFSYLANPGLHWKPILIQKVIPIDVS-TVREINTSGTILTYSEHFVQ 149
Query: 70 VVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQKNEIA---- 125
V ++QYR + K + ++N N ++ + +R+ + + N+D KNE +
Sbjct: 150 VNMTVQYRIVDPK--KYLFSVTNPDNCLRQSINSALRSVISRSNID--IFLKNEFSLLAK 205
Query: 126 KAVEEELEKAMSAYGYEIVQTLITDI 151
++ ++K + Y IV I+DI
Sbjct: 206 NDIKVNIQKIIKPYHMGIV---ISDI 228
>HFLK_HAEIN (P44546) HflK protein
Length = 410
Score = 40.4 bits (93), Expect = 0.005
Identities = 38/177 (21%), Positives = 80/177 (44%), Gaps = 6/177 (3%)
Query: 21 FGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCETKTKDNVFVNVVASIQYRALA 80
FG+ ++QPG + P F+ K + ++ +V++L + T+D V V ++QYR +
Sbjct: 118 FGELHSIVQPGLNWKPTFVDKVLPVNVE-QVKELRTQGAMLTQDENMVKVEMTVQYR-VQ 175
Query: 81 DKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ-KNEIAKAVEEELEKAMSAY 139
D A F ++N + + +R + ++++D ++ + + + L + + +Y
Sbjct: 176 DPAKYLF-SVTNADDSLNQATDSALRYVIGHMSMNDILTTGRSVVRENTWKALNEIIKSY 234
Query: 140 --GYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEAEKILQIKRAEGEAE 194
G E++ P VK A ++ A +AEA + A G+A+
Sbjct: 235 DMGLEVIDVNFQSARPPEEVKDAFDDAIKAQEDEQRFIREAEAYAREKEPIARGDAQ 291
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,311,214
Number of Sequences: 164201
Number of extensions: 1100240
Number of successful extensions: 4350
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 55
Number of HSP's that attempted gapping in prelim test: 4313
Number of HSP's gapped (non-prelim): 80
length of query: 286
length of database: 59,974,054
effective HSP length: 109
effective length of query: 177
effective length of database: 42,076,145
effective search space: 7447477665
effective search space used: 7447477665
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146776.3


