

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.17 - phase: 0
(304 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
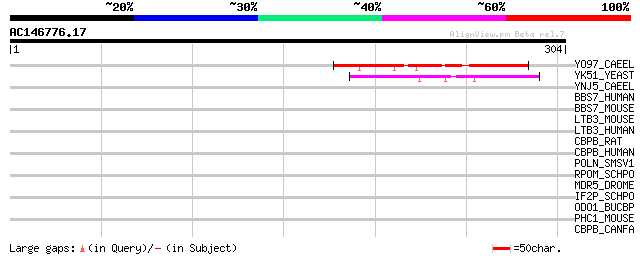
Score E
Sequences producing significant alignments: (bits) Value
YO97_CAEEL (P41847) Hypothetical protein T20B12.7 in chromosome III 108 2e-23
YK51_YEAST (P36152) Hypothetical 38.5 kDa protein in MET1-SIS2 i... 78 3e-14
YNJ5_CAEEL (P34549) Hypothetical protein R10E11.5 in chromosome III 34 0.39
BBS7_HUMAN (Q8IWZ6) Bardet-Biedl syndrome 7 protein (BBS2-like p... 34 0.39
BBS7_MOUSE (Q8K2G4) Bardet-Biedl syndrome 7 protein homolog (BBS... 33 1.1
LTB3_MOUSE (Q61810) Latent transforming growth factor beta bindi... 32 1.9
LTB3_HUMAN (Q9NS15) Latent transforming growth factor beta bindi... 31 3.3
CBPB_RAT (P19223) Carboxypeptidase B precursor (EC 3.4.17.2) 31 3.3
CBPB_HUMAN (P15086) Carboxypeptidase B precursor (EC 3.4.17.2) (... 31 3.3
POLN_SMSV1 (P36286) Non-structural polyprotein [Contains: Unknow... 31 4.3
RPOM_SCHPO (O13993) DNA-directed RNA polymerase, mitochondrial p... 30 5.7
MDR5_DROME (Q00748) Multidrug resistance protein homolog 65 (P-g... 30 5.7
IF2P_SCHPO (Q10251) Eukaryotic translation initiation factor 5B ... 30 5.7
ODO1_BUCBP (Q89AJ7) Oxoglutarate dehydrogenase (EC 1.2.4.2) 30 7.4
PHC1_MOUSE (Q64028) Polyhomeotic-like protein 1 (Early developme... 30 9.7
CBPB_CANFA (P55261) Carboxypeptidase B precursor (EC 3.4.17.2) (... 30 9.7
>YO97_CAEEL (P41847) Hypothetical protein T20B12.7 in chromosome III
Length = 238
Score = 108 bits (270), Expect = 2e-23
Identities = 62/119 (52%), Positives = 77/119 (64%), Gaps = 18/119 (15%)
Query: 178 KFVKSSPKVQIDF----DSDLIDKNSLLSEEDLKKP---ELPSGDCEIGPT-----RKAC 225
K V KV +D D DLID++ LL EED +KP +L +G C GP ++AC
Sbjct: 118 KLVNFGEKVALDLGVVADEDLIDEDGLLQEEDFEKPTGDQLKAGGC--GPDDPNKKKRAC 175
Query: 226 KNCSCGRAEEEEKVLKLGLTAEQINNPQSACGSCGLGDAFRCSTCPYKGLPAFKMGETV 284
KNCSCG AE+EE + K+G A + P+S+CG+C LGDAFRCSTCPY G P FK GETV
Sbjct: 176 KNCSCGLAEQEE-LEKMGQIAAE---PKSSCGNCSLGDAFRCSTCPYLGQPPFKPGETV 230
>YK51_YEAST (P36152) Hypothetical 38.5 kDa protein in MET1-SIS2
intergenic region
Length = 348
Score = 77.8 bits (190), Expect = 3e-14
Identities = 48/125 (38%), Positives = 64/125 (50%), Gaps = 23/125 (18%)
Query: 187 QIDFDSDLIDKNSLLSEEDLKKPELPSGDCEIGPTRK--ACKNCSCGRAEEEE------- 237
Q+D D I++ L+ E+ K + C T+K ACK+C+CG E+EE
Sbjct: 223 QVDTSDDSIEEEELIDEDGSGKSMITMITCGKSKTKKKKACKDCTCGMKEQEENEINDIR 282
Query: 238 ----KVLKLGLTAEQINNPQ--------SACGSCGLGDAFRCSTCPYKGLPAFKMGETVL 285
KV+K T +++ CGSC LGDAFRCS CPY GLPAFK G+ +
Sbjct: 283 SQQDKVVKF--TEDELTEIDFTIDGKKVGGCGSCSLGDAFRCSGCPYLGLPAFKPGQPIN 340
Query: 286 LLSPS 290
L S S
Sbjct: 341 LDSIS 345
>YNJ5_CAEEL (P34549) Hypothetical protein R10E11.5 in chromosome III
Length = 529
Score = 34.3 bits (77), Expect = 0.39
Identities = 31/113 (27%), Positives = 52/113 (45%), Gaps = 3/113 (2%)
Query: 32 DAAKMYGAVLACTDEAVLPVSQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSVD 91
D AKM GA+ +E V + Q I+ NE + ++ + ++K +E S+
Sbjct: 383 DVAKMSGALQKAEEEVVQTIDQTVKNIKSNVNEVKKDVEKNIAEKVDDITK-ELEKSAKS 441
Query: 92 LVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMIPDLENK 144
L K + ID +Q + L+ + KS AV SG+K+I DL ++
Sbjct: 442 LEETTDK-IGSKIDNTSQAIKSNLEE-ASLKTEKSVNDAVKSGEKLIGDLASE 492
>BBS7_HUMAN (Q8IWZ6) Bardet-Biedl syndrome 7 protein (BBS2-like
protein 1)
Length = 715
Score = 34.3 bits (77), Expect = 0.39
Identities = 40/150 (26%), Positives = 66/150 (43%), Gaps = 6/150 (4%)
Query: 63 NEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTL 122
NE V + D ++ S +S+ V S D +V+ + + LT E + G L
Sbjct: 275 NEPVLRFDQMLSESVTSIQGGCVGKDSYDEIVV--STYSGWVTGLTTEPIHKESGPGEEL 332
Query: 123 -IHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKKFVK 181
I++ Q+ + S + L+ K+L + Q+ QSS K+ PS+ I F L K
Sbjct: 333 KINQEMQNKISSLRNELEHLQYKVLQERENYQQSSQSSKAKSAVPSFGINDKFTLNKDDA 392
Query: 182 SSP---KVQIDFDSDLIDKNSLLSEEDLKK 208
S +VQ D+ LI + + D+ K
Sbjct: 393 SYSLILEVQTAIDNVLIQSDVPIDLLDVDK 422
>BBS7_MOUSE (Q8K2G4) Bardet-Biedl syndrome 7 protein homolog
(BBS2-like protein 1)
Length = 715
Score = 32.7 bits (73), Expect = 1.1
Identities = 34/120 (28%), Positives = 52/120 (43%), Gaps = 3/120 (2%)
Query: 60 ELGNEGVEKLDPLVITSASSLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGG 119
E NE V + D ++ S +S+ V D +VL S + LT E G
Sbjct: 272 ENANEPVLRFDQMLSESVTSIQGGCVGKDGYDEIVLATYS--GWVTGLTTEPTHKESGPG 329
Query: 120 TTL-IHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKK 178
L +++ Q+ + S I L+ K+L + Q+ QSS K+ PS+ I F L K
Sbjct: 330 EELKLNQEMQNKISSLRSEIEHLQFKVLQERENYQQSSQSSQAKSTVPSFSINDKFTLNK 389
>LTB3_MOUSE (Q61810) Latent transforming growth factor beta binding
protein 3 precursor (LTBP-3)
Length = 1268
Score = 32.0 bits (71), Expect = 1.9
Identities = 26/99 (26%), Positives = 34/99 (34%), Gaps = 23/99 (23%)
Query: 204 EDLKKPELPS----GDCEIGPTRKAC------------KNCSCGRAEEEEKVLKLGLTAE 247
+D+ + +P GDC P C C + EE+ +L T
Sbjct: 351 QDINECAMPGNVCHGDCLNNPGSYRCVCPPGHSLGPLAAQCIADKPEEKSLCFRLVSTEH 410
Query: 248 QINNP-------QSACGSCGLGDAFRCSTCPYKGLPAFK 279
Q +P Q C S G RC CP G AFK
Sbjct: 411 QCQHPLTTRLTRQLCCCSVGKAWGARCQRCPADGTAAFK 449
>LTB3_HUMAN (Q9NS15) Latent transforming growth factor beta binding
protein 3 precursor (LTBP-3)
Length = 1302
Score = 31.2 bits (69), Expect = 3.3
Identities = 21/69 (30%), Positives = 27/69 (38%), Gaps = 10/69 (14%)
Query: 218 IGPTRKACKNCSCGRAEEEEKVLKLGLTAEQINNP-------QSACGSCGLGDAFRCSTC 270
+GP+R C + EE+ +L Q +P Q C S G RC C
Sbjct: 386 LGPSRT---QCIADKPEEKSLCFRLVSPEHQCQHPLTTRLTRQLCCCSVGKAWGARCQRC 442
Query: 271 PYKGLPAFK 279
P G AFK
Sbjct: 443 PTDGTAAFK 451
>CBPB_RAT (P19223) Carboxypeptidase B precursor (EC 3.4.17.2)
Length = 415
Score = 31.2 bits (69), Expect = 3.3
Identities = 13/29 (44%), Positives = 16/29 (54%)
Query: 213 SGDCEIGPTRKACKNCSCGRAEEEEKVLK 241
+G CE+G +R C CG A E EK K
Sbjct: 254 AGWCEVGASRSPCSETYCGPAPESEKETK 282
>CBPB_HUMAN (P15086) Carboxypeptidase B precursor (EC 3.4.17.2)
(Pancreas-specific protein) (PASP)
Length = 417
Score = 31.2 bits (69), Expect = 3.3
Identities = 14/29 (48%), Positives = 16/29 (54%)
Query: 213 SGDCEIGPTRKACKNCSCGRAEEEEKVLK 241
+G CEIG +R C CG A E EK K
Sbjct: 256 AGWCEIGASRNPCDETYCGPAAESEKETK 284
>POLN_SMSV1 (P36286) Non-structural polyprotein [Contains: Unknown
protein; Helicase (2C-like protein) (P2C); 3A-like
protein; Viral genome-linked protein (VPg); Thiol
protease P3C (EC 3.4.22.-) (3C-like protease) (3C-pro);
RNA-directed RNA polymerase (EC
Length = 1879
Score = 30.8 bits (68), Expect = 4.3
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 10/68 (14%)
Query: 209 PELPSGDCEIGPTRKACKNCSCGRAEEEEKVLKL-----GLTAEQINNPQSACGSCGLGD 263
PE P + + G ++CK C+ A + + V GLT ++ AC SC L D
Sbjct: 98 PETPCCEAKDGGVVRSCKTCNLKPAHDSKAVSFFPAQTDGLTGDEPEFIAEACPSCVLYD 157
Query: 264 AFRCSTCP 271
TCP
Sbjct: 158 -----TCP 160
>RPOM_SCHPO (O13993) DNA-directed RNA polymerase, mitochondrial
precursor (EC 2.7.7.6)
Length = 1120
Score = 30.4 bits (67), Expect = 5.7
Identities = 29/126 (23%), Positives = 57/126 (45%), Gaps = 7/126 (5%)
Query: 89 SVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMI----PDLENK 144
S + V LI L+ PI+ T L++L +T++ +S +GD + PD+E
Sbjct: 151 SPEEVKLIMDQLNIPINNFTPSQLQLLGITNSTIVGESENGKDQNGDSSLKEKQPDVETT 210
Query: 145 LLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKKFVKSSPKVQIDFDSDLIDKNSLLSEE 204
+ + + + AL+SS+ S + +F + L++K+++LS
Sbjct: 211 VTKS--ANLNALRSSLSSLLTESIDLPIDEVSLEFGNQGDTFNL-ARQKLLEKSAILSAA 267
Query: 205 DLKKPE 210
++ K E
Sbjct: 268 EVWKSE 273
>MDR5_DROME (Q00748) Multidrug resistance protein homolog 65
(P-glycoprotein 65)
Length = 1302
Score = 30.4 bits (67), Expect = 5.7
Identities = 31/144 (21%), Positives = 68/144 (46%), Gaps = 16/144 (11%)
Query: 79 SLSKFPVESSSVDLVVLIWKSLDFPIDQLTQEVLRVLKAGGTTLIHKSSQSAVGSGDKMI 138
++++ +++ + L+ +LD+ ++ Q+ L + G TT++ SA+ DK++
Sbjct: 553 AIARALIQNPKILLLDEATSALDYQSEKQVQQALDLASKGRTTIVVSHRLSAIRGADKIV 612
Query: 139 PDLENKLLLAG-FSEIQALQS---SVIKA----------KKPSWKIGSSFALKKFVKSSP 184
+ K+L G ++ AL+ ++++A K+ S + +L F KS
Sbjct: 613 FIHDGKVLEEGSHDDLMALEGAYYNMVRAGDINMPDEVEKEDSIEDTKQKSLALFEKSFE 672
Query: 185 KVQIDFDSDLIDKNSLLSEEDLKK 208
++F+ KNS+ EE + K
Sbjct: 673 TSPLNFEKG--QKNSVQFEEPIIK 694
>IF2P_SCHPO (Q10251) Eukaryotic translation initiation factor 5B
(eIF-5B) (Translation initiation factor IF-2)
Length = 1079
Score = 30.4 bits (67), Expect = 5.7
Identities = 28/97 (28%), Positives = 44/97 (44%), Gaps = 6/97 (6%)
Query: 115 LKAGGTTLIHKSSQSAVGSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIG-SS 173
+KA G L K + + ++ LE+ + +AG S + Q V KK S + G SS
Sbjct: 307 MKAQGKYLSKKQKEQQALAQRRLQQMLESGVRVAGLSNGEKKQKPVYTNKKKSNRSGTSS 366
Query: 174 FALKKFVKSSPKVQIDFDSDLIDKNSLLSEEDLKKPE 210
+ ++SSP I D D S++D +K E
Sbjct: 367 ISSSGILESSPATSISVDEPQKD-----SKDDSEKVE 398
>ODO1_BUCBP (Q89AJ7) Oxoglutarate dehydrogenase (EC 1.2.4.2)
Length = 916
Score = 30.0 bits (66), Expect = 7.4
Identities = 23/70 (32%), Positives = 42/70 (59%), Gaps = 4/70 (5%)
Query: 52 SQVFDAIRELGNEGVEKLDPLVITSASSLSKFPVESSSV-DLVVLIWKSLDFPIDQL-TQ 109
SQ++ IR+ ++K PL+I S SL +FP+ +SS+ +L ++++ ID L T+
Sbjct: 737 SQIYHIIRKQAFSLIKK--PLIIMSPKSLLRFPLAASSLSELSNGKFRTVIDEIDNLDTK 794
Query: 110 EVLRVLKAGG 119
+V R++ G
Sbjct: 795 KVQRIILCSG 804
>PHC1_MOUSE (Q64028) Polyhomeotic-like protein 1 (Early development
regulator protein 1) (MPH1) (RAE-28)
Length = 1012
Score = 29.6 bits (65), Expect = 9.7
Identities = 22/95 (23%), Positives = 41/95 (43%), Gaps = 7/95 (7%)
Query: 132 GSGDKMIPDLENKLLLAGFSEIQALQSSVIKAKKPSWKIGSSFALKKFVKSSPKVQIDFD 191
G G++ DL N + E+Q + + + W + + ++ ++ +F
Sbjct: 915 GHGER---DLGNTITTPSTPELQGINPVFLSSNPSQWSVEEVYEFIASLQGCQEIAEEFR 971
Query: 192 SDLIDKNSLLSEEDLKKPELPSG-DCEIGPTRKAC 225
S ID +LL LK+ L S + ++GP K C
Sbjct: 972 SQEIDGQALLL---LKEEHLMSAMNIKLGPALKIC 1003
>CBPB_CANFA (P55261) Carboxypeptidase B precursor (EC 3.4.17.2) (47
kDa zymogen granule membrane associated protein) (ZAP47)
Length = 416
Score = 29.6 bits (65), Expect = 9.7
Identities = 13/29 (44%), Positives = 16/29 (54%)
Query: 213 SGDCEIGPTRKACKNCSCGRAEEEEKVLK 241
+G C+IG +R C CG A E EK K
Sbjct: 255 AGWCKIGASRNPCDETYCGPAAESEKETK 283
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,699,318
Number of Sequences: 164201
Number of extensions: 1346918
Number of successful extensions: 3944
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 3930
Number of HSP's gapped (non-prelim): 19
length of query: 304
length of database: 59,974,054
effective HSP length: 110
effective length of query: 194
effective length of database: 41,911,944
effective search space: 8130917136
effective search space used: 8130917136
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146776.17


