

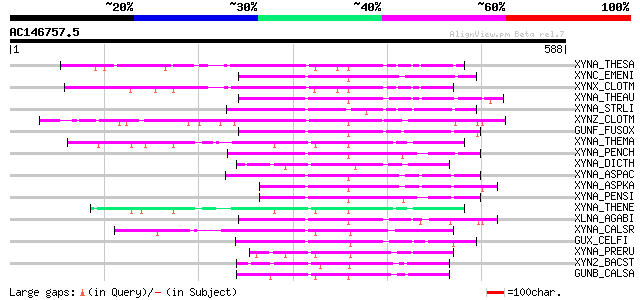
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.5 - phase: 0 /pseudo
(588 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
XYNA_THESA (P36917) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 114 7e-25
XYNC_EMENI (Q00177) Endo-1,4-beta-xylanase C precursor (EC 3.2.1... 110 1e-23
XYNX_CLOTM (P38535) Exoglucanase xynX precursor (EC 3.2.1.91) (E... 107 7e-23
XYNA_THEAU (P23360) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 105 3e-22
XYNA_STRLI (P26514) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 105 4e-22
XYNZ_CLOTM (P10478) Endo-1,4-beta-xylanase Z precursor (EC 3.2.1... 104 7e-22
GUNF_FUSOX (P46239) Putative endoglucanase type F precursor (EC ... 101 6e-21
XYNA_THEMA (Q60037) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 98 5e-20
XYNA_PENCH (P29417) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 97 9e-20
XYNA_DICTH (Q12603) Beta-1,4-xylanase (EC 3.2.1.8) (Endo-1,4-bet... 94 1e-18
XYNA_ASPAC (O59859) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 93 2e-18
XYNA_ASPKA (P33559) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 92 5e-18
XYNA_PENSI (P56588) Endo-1,4-beta-xylanase (EC 3.2.1.8) (Xylanas... 91 6e-18
XYNA_THENE (Q60042) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 91 1e-17
XLNA_AGABI (O60206) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 91 1e-17
XYNA_CALSR (P40944) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 90 2e-17
GUX_CELFI (P07986) Exoglucanase/xylanase precursor [Includes: Ex... 86 2e-16
XYNA_PRERU (P48789) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 85 6e-16
XYN2_BACST (P45703) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 84 8e-16
GUNB_CALSA (P10474) Endoglucanase/exoglucanase B precursor [Incl... 84 1e-15
>XYNA_THESA (P36917) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 1157
Score = 114 bits (285), Expect = 7e-25
Identities = 116/467 (24%), Positives = 215/467 (45%), Gaps = 75/467 (16%)
Query: 54 QKPQYNGGIIKNPELNDG-LQGWTTFGDAIIEHRKSL---GNKFVVTHSR----NQPHDS 105
Q P G +I N +G GW G ++++ + G+ ++T R N P
Sbjct: 189 QNPIQVGNVIANETFENGNTSGWIGTGSSVVKAVYGVAHSGDYSLLTTGRTANWNGPSYD 248
Query: 106 VSQKIYLRKGLHYSLSAWIQV--SEETVPVTAVVKTT--KGFKFGGAIFAEPNC--WSML 159
++ KI G Y++ W++ +T + A VK T K FA N W+ +
Sbjct: 249 LTGKIV--PGQQYNVDFWVKFVNGNDTEQIKATVKATSDKDNYIQVNDFANVNKGEWTEI 306
Query: 160 KGGL---IADTTGVAELYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVV 216
KG +AD +G++ +Y ES N ++E ++D+ S+ S+ +++I+ D
Sbjct: 307 KGSFTLPVADYSGIS-IYVESQNPTLEFYIDDFSVIGEI-----SNNQITIQND------ 354
Query: 217 VRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNN-NAYQDWFASRFTVTTFENEMK 275
+P+ ++ + FP G A++ + LN+ + + A F + EN MK
Sbjct: 355 ----------IPDL---YSVFKDYFPIGVAVDPSRLNDADPHAQLTAKHFNMLVAENAMK 401
Query: 276 WYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWV--------SSLSP 327
+ + +G + +AD ++ +A + +RGH + W + P+W S S
Sbjct: 402 PESLQPTEGNFTFDNADKIVDYAIAHNMKMRGHTLLWHN--QVPDWFFQDPSDPSKSASR 459
Query: 328 DQLNDAVEKRVNSIVSRYK------GQLIGWDVVNE------NLHFSFFESKLGQNFSAR 375
D L ++ + +++ +K +IGWDVVNE NL S + +G ++ +
Sbjct: 460 DLLLQRLKTHITTVLDHFKTKYGSQNPIIGWDVVNEVLDDNGNLRNSKWLQIIGPDYIEK 519
Query: 376 MFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFP 435
F H D LF+N+YN +G+ T Y + +K+++S + GIG++ H
Sbjct: 520 AFEYAHEADPSMKLFINDYNI---ENNGVKTQAMY-DLVKKLKSEGVPID-GIGMQMHI- 573
Query: 436 NSPPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQVLREA 482
N N+ ++AS++ L + G+ I +TELD+ N + E +L++A
Sbjct: 574 NINSNIDNIKASIEKLASLGVEIQVTELDMNMNGN--ISNEALLKQA 618
>XYNC_EMENI (Q00177) Endo-1,4-beta-xylanase C precursor (EC 3.2.1.8)
(Xylanase C) (1,4-beta-D-xylan xylanohydrolase C) (34
kDa xylanase) (Xylanase X34)
Length = 327
Score = 110 bits (274), Expect = 1e-23
Identities = 77/261 (29%), Positives = 127/261 (48%), Gaps = 18/261 (6%)
Query: 243 FGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQG 302
FG+ ++ +L N+ + AS+F V T EN MKW E +QG + AD ++ +A +
Sbjct: 41 FGTCSDQALLQNSQNEAIVASQFGVITPENSMKWDALEPSQGNFGWSGADYLVDYATQHN 100
Query: 303 IAVRGHNIFWDDPQYQPNWVSSL-SPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE---- 357
VRGH + W P+WVSS+ + L + +N +V RYKG+++ WDVVNE
Sbjct: 101 KKVRGHTLVWH--SQLPSWVSSIGDANTLRSVMTNHINEVVGRYKGKIMHWDVVNEIFNE 158
Query: 358 --NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIK 415
S F + LG++F F D L++N+YN S +Y++K
Sbjct: 159 DGTFRNSVFYNLLGEDFVRIAFETARAADPDAKLYINDYNLDSASYAKTQAMASYVKK-- 216
Query: 416 EIQSVNSQLPL-GIGLESHFPNSPPNLPYMRASLDTLRATGL-PIWITELDVASQPNQAL 473
+ +P+ GIG ++H+ +S + +L +L TG+ + ITELD+A +
Sbjct: 217 ---WLAEGVPIDGIGSQAHYSSSHWSSTEAAGALSSLANTGVSEVAITELDIAGAASSD- 272
Query: 474 YFEQVLREAHSHPGIQGIVMW 494
+ +L + GI +W
Sbjct: 273 -YLNLLNACLNEQKCVGITVW 292
>XYNX_CLOTM (P38535) Exoglucanase xynX precursor (EC 3.2.1.91)
(Exocellobiohydrolase) (1,4-beta-cellobiohydrolase)
Length = 1087
Score = 107 bits (268), Expect = 7e-23
Identities = 101/452 (22%), Positives = 204/452 (44%), Gaps = 67/452 (14%)
Query: 59 NGGIIKNPELNDG-LQGWTTFGDAIIEH--RKSLGNKFVVTHSRNQPHDSVSQKIY--LR 113
N ++ N + G + GW G+ + +++G + R Q ++ + ++
Sbjct: 36 NANLVSNGDFESGTIDGWIKQGNPTLAATTEEAIGQYSMKVAGRTQTYEGPAYSFLGKMQ 95
Query: 114 KGLHYSLSAWIQV------SEETVPVTAVVKTTKGFKFGGAIFAEP---NCWSMLKGGLI 164
KG Y++S +++ S + VT + G + ++ + + W+ + G
Sbjct: 96 KGQSYNVSLKVRLVSGQNSSNPLITVTMFREDDNGKHYDTIVWQKQVSEDSWTTVNGTYT 155
Query: 165 ADTTGVAE---LYFESNNTSVEIWVDNVSLQPFTEKQWR--SHQELSIEKDRKRKVVVRA 219
D TG + +Y ES + ++E ++D+V + P Q S+ +++I+ D
Sbjct: 156 LDYTGTLKTLYMYVESPDPTLEYYIDDVVVTPQNPIQVGEISNNQITIQND--------- 206
Query: 220 VNEQGHPLPNASISLTMKRPGFPFGSAINKNILNN-NAYQDWFASRFTVTTFENEMKWYT 278
+P+ S ++ + FP G A++ + LN+ + + A F + EN MK +
Sbjct: 207 -------IPDLS---SVFKDYFPIGVAVDPSRLNDTDPHAQLTAKHFNMLVAENAMKPES 256
Query: 279 NEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNW--------VSSLSPDQL 330
+ +G + +AD ++ +A + +RGH + W + P+W S D L
Sbjct: 257 LQPTEGNFTFDNADRIVDYAIAHNMKMRGHTLLWHN--QVPDWFFQDPSDPTKPASRDLL 314
Query: 331 NDAVEKRVNSIVSRYK------GQLIGWDVVNE------NLHFSFFESKLGQNFSARMFN 378
++ + +++ +K +IGWDVVNE +L S + +G ++ + F
Sbjct: 315 LQRLKTHITTVLDHFKTKYGAQNPIIGWDVVNEVLDDNGSLRNSKWLQIIGPDYIEKAFE 374
Query: 379 EVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSP 438
H D LF+N+YN +G+ T Y + +K+++S + GIG++ H N
Sbjct: 375 YAHEADPSMKLFINDYNI---ENNGVKTQAMY-DLVKKLKSEGVPIS-GIGMQMHI-NIN 428
Query: 439 PNLPYMRASLDTLRATGLPIWITELDVASQPN 470
N+ ++AS++ L + G+ I +TELD+ N
Sbjct: 429 SNIDNIKASIEKLASLGVEIQVTELDMNMNGN 460
>XYNA_THEAU (P23360) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase) (TAXI)
Length = 329
Score = 105 bits (262), Expect = 3e-22
Identities = 84/296 (28%), Positives = 141/296 (47%), Gaps = 29/296 (9%)
Query: 243 FGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQG 302
FG A ++N L + F T EN MKW E +QG N+ AD ++ +A++ G
Sbjct: 44 FGVATDQNRLTTGKNAAIIQADFGQVTPENSMKWDATEPSQGNFNFAGADYLVNWAQQNG 103
Query: 303 IAVRGHNIFWDDPQYQPNWVSSLS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE---- 357
+RGH + W P+WVSS++ + L + ++ + ++++RYKG++ WDVVNE
Sbjct: 104 KLIRGHTLVWH--SQLPSWVSSITDKNTLTNVMKNHITTLMTRYKGKIRAWDVVNEAFNE 161
Query: 358 --NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPT--YIEK 413
+L + F + +G+++ F D L++N+YN D S P T + +
Sbjct: 162 DGSLRQTVFLNVIGEDYIPIAFQTARAADPNAKLYINDYNL-----DSASYPKTQAIVNR 216
Query: 414 IKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLP-IWITELDVA-SQPNQ 471
+K+ ++ + GIG ++H S + +L L + G P + ITELDVA + P
Sbjct: 217 VKQWRAAGVPID-GIGSQTHL--SAGQGAGVLQALPLLASAGTPEVAITELDVAGASPTD 273
Query: 472 ALYFEQVLREAHSHPGIQGIVMWTAWSPQGCYRICLT----DNNFKNLPAGDVVDQ 523
+ V+ + GI +W P +R T D NF PA + + Q
Sbjct: 274 ---YVNVVNACLNVQSCVGITVWGVADPDS-WRASTTPLLFDGNFNPKPAYNAIVQ 325
>XYNA_STRLI (P26514) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 477
Score = 105 bits (261), Expect = 4e-22
Identities = 74/276 (26%), Positives = 122/276 (43%), Gaps = 23/276 (8%)
Query: 230 ASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYF 289
+++ + G FG+AI L+++ Y F + T ENEMK E +G+ N+
Sbjct: 44 STLGAAAAQSGRYFGTAIASGRLSDSTYTSIAGREFNMVTAENEMKIDATEPQRGQFNFS 103
Query: 290 DADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQL 349
AD + +A + G VRGH + W QP W+ SLS L A+ +N +++ YKG++
Sbjct: 104 SADRVYNWAVQNGKQVRGHTLAWH--SQQPGWMQSLSGSALRQAMIDHINGVMAHYKGKI 161
Query: 350 IGWDVVNENLHFSFFESKLGQNFSARM-----------FNEVHNIDGQTTLFMNEYNTIE 398
+ WDVVNE +F + G + + F D L N+YN
Sbjct: 162 VQWDVVNE----AFADGSSGARRDSNLQRSGNDWIEVAFRTARAADPSAKLCYNDYNV-- 215
Query: 399 DSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPI 458
++ T Y +++ + + +G +SHF + P R +L A G+ +
Sbjct: 216 ENWTWAKTQAMY-NMVRDFKQRGVPIDC-VGFQSHFNSGSPYNSNFRTTLQNFAALGVDV 273
Query: 459 WITELDVASQPNQALYFEQVLREAHSHPGIQGIVMW 494
ITELD+ P A + V + + GI +W
Sbjct: 274 AITELDIQGAP--ASTYANVTNDCLAVSRCLGITVW 307
>XYNZ_CLOTM (P10478) Endo-1,4-beta-xylanase Z precursor (EC 3.2.1.8)
(Xylanase Z) (1,4-beta-D-xylan xylanohydrolase Z)
Length = 837
Score = 104 bits (259), Expect = 7e-22
Identities = 127/563 (22%), Positives = 232/563 (40%), Gaps = 96/563 (17%)
Query: 32 TGFEAEALSYDYSANVECLAHPQKPQYNGGIIKNPELNDGLQGWTTFGDAIIEHRKSLGN 91
T EAE S+++E + P PE G+ G+ T GD ++ GN
Sbjct: 299 TRIEAEDYDGINSSSIEIIGVP------------PEGGRGI-GYITSGDYLVYKSIDFGN 345
Query: 92 KFVVTHSRNQPHDSVSQKIYLRKG-----LHYSLSA-----WIQVSEETVPVTAVVKTTK 141
T + + ++ + I LR L +LS W E+T ++ V
Sbjct: 346 G--ATSFKAKVANANTSNIELRLNGPNGTLIGTLSVKSTGDWNTYEEQTCSISKVT---- 399
Query: 142 GFKFGGAIFAEPNCWSMLKGGLIADTTGVAELYFESNNTSVEIWVDN---VSLQPFTEKQ 198
G +F P G+ + +TG+ +L + N S ++ + + P T +
Sbjct: 400 GINDLYLVFKGPVNIDWFTFGVESSSTGLGDLNGDGNINSSDLQALKRHLLGISPLTGEA 459
Query: 199 W------RSHQELSIEKDRKRKVVVRAVNE---QGH-PLPNASISLTM------------ 236
RS + S + ++ ++R + E QG PN S++ T
Sbjct: 460 LLRADVNRSGKVDSTDYSVLKRYILRIITEFPGQGDVQTPNPSVTPTQTPIPTISGNALR 519
Query: 237 ---KRPGFPFGSAINKNILNNN--AYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDA 291
+ G G+ +N NN+ Y F++ ENEMK+ + Q ++
Sbjct: 520 DYAEARGIKIGTCVNYPFYNNSDPTYNSILQREFSMVVCENEMKFDALQPRQNVFDFSKG 579
Query: 292 DAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVS--SLSPDQLNDAVEKRVNSIVSRYKGQL 349
D +L FAE+ G+ +RGH + W + P+W++ + + D L ++ + ++++ YKG++
Sbjct: 580 DQLLAFAERNGMQMRGHTLIWHN--QNPSWLTNGNWNRDSLLAVMKNHITTVMTHYKGKI 637
Query: 350 IGWDVVNE-------NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRD 402
+ WDV NE L S + + +GQ++ F D LF N+YN IED
Sbjct: 638 VEWDVANECMDDSGNGLRSSIWRNVIGQDYLDYAFRYAREADPDALLFYNDYN-IEDLGP 696
Query: 403 GLSTPPTYIEKIKEIQSVNSQLPL-GIGLESHFPN--SPPNLPYMRASLDTLRATGLPIW 459
+ I+ +KE +P+ G+G + HF N SP L + ++ G+ +
Sbjct: 697 KSNAVFNMIKSMKE-----RGVPIDGVGFQCHFINGMSPEYLASIDQNIKRYAEIGVIVS 751
Query: 460 ITELDVASQPN---------QALYFEQVLREAHSHPGIQGIVMW-----TAWSP---QGC 502
TE+D+ + QA ++++++ ++P VMW W P G
Sbjct: 752 FTEIDIRIPQSENPATAFQVQANNYKELMKICLANPNCNTFVMWGFTDKYTWIPGTFPGY 811
Query: 503 YRICLTDNNFKNLPAGDVVDQLI 525
+ D+N+ PA + + + +
Sbjct: 812 GNPLIYDSNYNPKPAYNAIKEAL 834
>GUNF_FUSOX (P46239) Putative endoglucanase type F precursor (EC
3.2.1.4) (Endo-1,4-beta-glucanase) (Cellulase)
Length = 385
Score = 101 bits (251), Expect = 6e-21
Identities = 80/274 (29%), Positives = 127/274 (46%), Gaps = 25/274 (9%)
Query: 243 FGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQG 302
FG+ I+ LNNN + ++F T EN MKW E ++ + +AD ++ FA + G
Sbjct: 96 FGTEIDHYHLNNNPLINIVKAQFGQVTCENSMKWDAIEPSRNSFTFSNADKVVDFATQNG 155
Query: 303 IAVRGHNIFWDDPQYQPNWVSSLSP-DQLNDAVEKRVNSIVSRYKGQLIGWDVVNE---- 357
+RGH + W P WV +++ L +E V ++V+RYKG+++ WDVVN
Sbjct: 156 KLIRGHTLLWH--SQLPQWVQNINDRSTLTAVIENHVKTMVTRYKGKILQWDVVNNEIFA 213
Query: 358 ---NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKI 414
NL S F LG++F F D L++N+YN D D + +
Sbjct: 214 EDGNLRDSVFSRVLGEDFVGIAFRAARAADPAAKLYINDYNL--DKSDYAKVTRGMVAHV 271
Query: 415 KEIQSVNSQLPL-GIGLESHF--PNSPPNLPYMRASLDTLRAT-GLPIWITELDVASQPN 470
+ + + +P+ GIG + H P+ + A+L L A+ I ITELD+A
Sbjct: 272 N--KWIAAGIPIDGIGSQGHLAAPSGWNPASGVPAALRALAASDAKEIAITELDIAGA-- 327
Query: 471 QALYFEQVLREAHSHPGIQGIVMW-----TAWSP 499
A + V+ + P GI +W +W P
Sbjct: 328 SANDYLTVMNACLAVPKCVGITVWGVSDKDSWRP 361
>XYNA_THEMA (Q60037) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 1059
Score = 98.2 bits (243), Expect = 5e-20
Identities = 100/454 (22%), Positives = 185/454 (40%), Gaps = 63/454 (13%)
Query: 62 IIKNPELNDGLQGWTTFGDAIIEHRKSLGNK-----FVVTHSRNQPHDSVSQKIYLRKGL 116
+I +G+ W GD IE + + F+ + ++ K L+ G
Sbjct: 205 VIYETSFENGVGDWQPRGDVNIEASSEVAHSGKSSLFISNRQKGWQGAQINLKGILKTGK 264
Query: 117 HYSLSAWIQVS---EETVPVTAVVKTTKG----FKFGGAIFAEPNCWSMLKGGLIADTTG 169
Y+ AW+ + ++T+ +T K + +++ + W L G
Sbjct: 265 TYAFEAWVYQNSGQDQTIIMTMQRKYSSDASTQYEWIKSATVPSGQWVQLSGTYTIPAGV 324
Query: 170 VAE---LYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQGHP 226
E LYFES N ++E +VD+V + T E+ IE + ++++ A+ E
Sbjct: 325 TVEDLTLYFESQNPTLEFYVDDVKIVDTTSA------EIKIEMEPEKEIP--ALKE---- 372
Query: 227 LPNASISLTMKRPGFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTN----EYA 282
+ + F G A+ + N + F T ENEMK + E
Sbjct: 373 ---------VLKDYFKVGVALPSKVFLNPKDIELITKHFNSITAENEMKPESLLAGIENG 423
Query: 283 QGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWV------SSLSPDQLNDAVEK 336
+ K + AD + F E+ G+ +RGH + W + P+W + LS + + + +++
Sbjct: 424 KLKFRFETADKYIQFVEENGMVIRGHTLVWHNQT--PDWFFKDENGNLLSKEAMTERLKE 481
Query: 337 RVNSIVSRYKGQLIGWDVVNE--------NLHFSFFESKLGQNFSARMFNEVHNIDGQTT 388
++++V +KG++ WDVVNE L S + +G ++ F D
Sbjct: 482 YIHTVVGHFKGKVYAWDVVNEAVDPNQPDGLRRSTWYQIMGPDYIELAFKFAREADPDAK 541
Query: 389 LFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASL 448
LF N+YNT E + + ++ +KE ++ GIG++ H + A
Sbjct: 542 LFYNDYNTFEPRKRDIIY--NLVKDLKEKGLID-----GIGMQCHISLATDIKQIEEAIK 594
Query: 449 DTLRATGLPIWITELDVASQPNQALYFEQVLREA 482
G+ I ITELD++ + + + + R A
Sbjct: 595 KFSTIPGIEIHITELDMSVYRDSSSNYPEAPRTA 628
>XYNA_PENCH (P29417) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase)
Length = 353
Score = 97.4 bits (241), Expect = 9e-20
Identities = 77/283 (27%), Positives = 125/283 (43%), Gaps = 28/283 (9%)
Query: 231 SISLTMKRPGFPF-GSAINKNILNNNAYQDWFA-SRFTVTTFENEMKWYTNEYAQGKDNY 288
SI K G + G+ ++ LN N + F + EN MKW E +QG+ ++
Sbjct: 34 SIDAKFKAHGKKYLGNIADQGTLNGNPKTPAIIKANFGQLSPENSMKWDATEPSQGQFSF 93
Query: 289 FDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSLS-PDQLNDAVEKRVNSIVSRYKG 347
+D + FAE G +RGH + W P+WVSS++ L D ++ + +++ +YKG
Sbjct: 94 AGSDYFVEFAETNGKLIRGHTLVWH--SQLPSWVSSITDKTTLTDVMKNHITTVMKQYKG 151
Query: 348 QLIGWDVVNE------NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSR 401
+L WDVVNE L S F LG++F F D + L++N+YN +
Sbjct: 152 KLYAWDVVNEIFEEDGTLRDSVFSRVLGEDFVRIAFETAREADPEAKLYINDYNLDSATS 211
Query: 402 DGLSTPPTYIEKI----KEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGL- 456
L ++++K I + SQ LG G + +L+ L + G
Sbjct: 212 AKLQGMVSHVKKWIAAGVPIDGIGSQTHLGAGAGA----------AASGALNALASAGTE 261
Query: 457 PIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSP 499
+ +TELD+A + + V+ P GI +W P
Sbjct: 262 EVAVTELDIAGATSTD--YVDVVNACLDQPKCVGITVWGVADP 302
>XYNA_DICTH (Q12603) Beta-1,4-xylanase (EC 3.2.1.8)
(Endo-1,4-beta-xylanase) (1,4-beta-D-xylan
xylanohydrolase)
Length = 352
Score = 94.0 bits (232), Expect = 1e-18
Identities = 67/240 (27%), Positives = 113/240 (46%), Gaps = 30/240 (12%)
Query: 241 FPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFD---ADAMLGF 297
F G+A++ LN Y++ F T EN+MKW E K +D AD ++ F
Sbjct: 43 FTIGAAVSH--LNIYHYENLLKKHFNSLTPENQMKW---EVIHPKPYVYDFGPADEIVDF 97
Query: 298 AEKQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE 357
A K G+ VRGH + W + P WV + + D++ +++ + +V YKG++ WDVVNE
Sbjct: 98 AMKNGMKVRGHTLVWHNQT--PGWVYAGTKDEILARLKEHIKEVVGHYKGKVYAWDVVNE 155
Query: 358 NLHFSFFE--------SKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPT 409
L + E G+ + F H +D LF N+YN D +
Sbjct: 156 ALSDNPNEFLRRAPWYDICGEEVIEKAFIWAHEVDPDAKLFYNDYNL----EDPIKREKA 211
Query: 410 Y--IEKIKEIQSVNSQLPL-GIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVA 466
Y ++K+K+ +P+ GIG++ H+ + P + S+ G+ + +TE D++
Sbjct: 212 YKLVKKLKD-----KGVPIHGIGIQGHWTLAWPTPKMLEDSIKRFAELGVEVQVTEFDIS 266
>XYNA_ASPAC (O59859) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase)
(FIA-xylanase)
Length = 327
Score = 93.2 bits (230), Expect = 2e-18
Identities = 76/282 (26%), Positives = 131/282 (45%), Gaps = 22/282 (7%)
Query: 229 NASISLTMKRPGFPF-GSAINKNILNNNAYQDWFASR-FTVTTFENEMKWYTNEYAQGKD 286
+ SI K G + G+ ++ LN NA F T EN MKW E +G+
Sbjct: 28 SVSIDAKFKAHGKKYLGTIGDQYTLNKNAKTPAIIKADFGQLTPENSMKWDATEPNRGQF 87
Query: 287 NYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSSL-SPDQLNDAVEKRVNSIVSRY 345
++ +D ++ FA+ G +RGH + W P+WV S+ L ++ + +++ RY
Sbjct: 88 SFSGSDYLVNFAQSNGKLIRGHTLVWH--SQLPSWVQSIYDKGTLIQVMQNHIATVMQRY 145
Query: 346 KGQLIGWDVVNE------NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIED 399
KG++ WDVVNE +L S F + +G+++ F +D L++N+YN
Sbjct: 146 KGKVYAWDVVNEIFNEDGSLRQSHFYNVIGEDYVRIAFETARAVDPNAKLYINDYNLDSA 205
Query: 400 SRDGLSTPPTYIEKIKEIQSVNSQLPL-GIGLESHFPNSPPNLPYMRASLDTLRATGL-P 457
S L+ +++K V + +P+ GIG ++H S + +L+ L G
Sbjct: 206 SYPKLTGLVNHVKK-----WVAAGVPIDGIGSQTHL--SAGAGAAVSGALNALAGAGTKE 258
Query: 458 IWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWTAWSP 499
+ ITELD+A + + V++ + P GI +W P
Sbjct: 259 VAITELDIAGA--SSTDYVNVVKACLNQPKCVGITVWGVADP 298
>XYNA_ASPKA (P33559) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 327
Score = 91.7 bits (226), Expect = 5e-18
Identities = 68/265 (25%), Positives = 124/265 (46%), Gaps = 23/265 (8%)
Query: 265 FTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSS 324
F T EN MKW E ++G+ ++ +D ++ FA+ +RGH + W P+WV +
Sbjct: 66 FGALTPENSMKWDATEPSRGQFSFSGSDYLVNFAQSNNKLIRGHTLVWH--SQLPSWVQA 123
Query: 325 LS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE------NLHFSFFESKLGQNFSARMF 377
++ + L + ++ + +++ YKG++ WDVVNE +L S F +G ++ F
Sbjct: 124 ITDKNTLIEVMKNHITTVMQHYKGKIYAWDVVNEIFNEDGSLRDSVFYKVIGDDYVRIAF 183
Query: 378 NEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPL-GIGLESHFPN 436
D L++N+YN S L+ ++++K + + +P+ GIG ++H
Sbjct: 184 ETARAADPNAKLYINDYNLDSASYPKLAGMVSHVKK-----WIEAGIPIDGIGSQTHL-- 236
Query: 437 SPPNLPYMRASLDTLRATGL-PIWITELDVASQPNQALYFEQVLREAHSHPGIQGIVMWT 495
S + +L+ L G I +TELD+A + + +V+ P GI +W
Sbjct: 237 SAGGGAGISGALNALAGAGTKEIAVTELDIAGA--SSTDYVEVVEACLDQPKCIGITVWG 294
Query: 496 AWSP---QGCYRICLTDNNFKNLPA 517
P + L D+N+ PA
Sbjct: 295 VADPDSWRSSSTPLLFDSNYNPKPA 319
>XYNA_PENSI (P56588) Endo-1,4-beta-xylanase (EC 3.2.1.8) (Xylanase)
(1,4-beta-D-xylan xylanohydrolase)
Length = 302
Score = 91.3 bits (225), Expect = 6e-18
Identities = 65/247 (26%), Positives = 115/247 (46%), Gaps = 26/247 (10%)
Query: 265 FTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWVSS 324
F T EN MKW E +G+ + +D ++ FA+ G +RGH + W P WVSS
Sbjct: 41 FGQLTPENSMKWDATEPNRGQFTFSGSDYLVNFAQSNGKLIRGHTLVWH--SQLPGWVSS 98
Query: 325 LS-PDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE------NLHFSFFESKLGQNFSARMF 377
++ + L ++ + ++++RYKG++ WDV+NE +L S F + +G+++ F
Sbjct: 99 ITDKNTLISVLKNHITTVMTRYKGKIYAWDVLNEIFNEDGSLRNSVFYNVIGEDYVRIAF 158
Query: 378 NEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIK----EIQSVNSQLPLGIGLESH 433
++D L++N+YN ++ ++++K I + SQ LG G S
Sbjct: 159 ETARSVDPNAKLYINDYNLDSAGYSKVNGMVSHVKKWLAAGIPIDGIGSQTHLGAGAGS- 217
Query: 434 FPNSPPNLPYMRASLDTLRATGL-PIWITELDVASQPNQALYFEQVLREAHSHPGIQGIV 492
+ +L+ L + G I ITELD+A + + V+ + GI
Sbjct: 218 ---------AVAGALNALASAGTKEIAITELDIAGA--SSTDYVNVVNACLNQAKCVGIT 266
Query: 493 MWTAWSP 499
+W P
Sbjct: 267 VWGVADP 273
>XYNA_THENE (Q60042) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
(Endoxylanase)
Length = 1055
Score = 90.5 bits (223), Expect = 1e-17
Identities = 96/425 (22%), Positives = 172/425 (39%), Gaps = 61/425 (14%)
Query: 86 RKSLGNKFVVTHSRNQPHDSVSQKIYLRKGLHYSLSAWIQVS---EETVPVTAVVK---- 138
+KSL FV + +S K L+ G Y+ AW+ ++T+ +T K
Sbjct: 233 KKSL---FVSNRQKGWHGAQISLKGILKTGKTYAFEAWVYQESGQDQTIIMTMQRKYSSD 289
Query: 139 TTKGFKFGGAIFAEPNCWSMLKGGLIADTTGVAE---LYFESNNTSVEIWVDNVSLQPFT 195
++ +++ A W L G E LYFES N ++E +VD+V + T
Sbjct: 290 SSTKYEWIKAATVPSGQWVQLSGTYTIPAGVTVEDLTLYFESQNPTLEFYVDDVKVVDTT 349
Query: 196 EKQWRSHQELSIEKDRKRKVVVRAVNEQGHPLPNASISLTMKRPGFPFGSAINKNILNNN 255
E+ +E + + ++ + + F G A+ + N
Sbjct: 350 SA------EIKLEMNPEEEIPALK---------------DVLKDYFRVGVALPSKVFINQ 388
Query: 256 AYQDWFASRFTVTTFENEMKWYTN----EYAQGKDNYFDADAMLGFAEKQGIAVRGHNIF 311
+ +T ENEMK + E + K + AD + FA++ G+ VRGH +
Sbjct: 389 KDIALISKHSNSSTAENEMKPDSLLAGIENGKLKFRFETADKYIEFAQQNGMVVRGHTLV 448
Query: 312 WDDPQYQPNWV------SSLSPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNE-------- 357
W + P W + LS +++ + + + ++++V +KG++ WDVVNE
Sbjct: 449 WHNQT--PEWFFKDENGNLLSKEEMTERLREYIHTVVGHFKGKVYAWDVVNEAVDPNQPD 506
Query: 358 NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEI 417
L S + +G ++ F D LF N+YNT E + + ++ +KE
Sbjct: 507 GLRRSTWYQIMGPDYIELAFKFAREADPNAKLFYNDYNTFEPKKRDIIY--NLVKSLKEK 564
Query: 418 QSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQPNQALYFEQ 477
++ GIG++ H + A G+ I ITELD++ + + +
Sbjct: 565 GLID-----GIGMQCHISLATDIRQIEEAIKKFSTIPGIEIHITELDISVYRDSTSNYSE 619
Query: 478 VLREA 482
R A
Sbjct: 620 APRTA 624
>XLNA_AGABI (O60206) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase)
Length = 333
Score = 90.5 bits (223), Expect = 1e-17
Identities = 82/307 (26%), Positives = 128/307 (40%), Gaps = 46/307 (14%)
Query: 243 FGSAINKNILNNNAY--QDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEK 300
FG+A + L + Y Q + F T N MKW E ++G + + D + A
Sbjct: 31 FGTATDNPELGDAPYVAQLGNTADFNQITAGNSMKWDATEPSRGTFTFSNGDTVANMARN 90
Query: 301 QGIAVRGHNIFWDDPQYQPNWVSSLSPDQ--LNDAVEKRVNSIVSRYKGQLIGWDVVNE- 357
+G +RGH W PNWV+S + D L V+ +++VS Y+GQ+ WDVVNE
Sbjct: 91 RGQLLRGHTCVWH--SQLPNWVTSGNFDNSTLLSIVQNHCSTLVSHYRGQMYSWDVVNEP 148
Query: 358 -----NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPT-YI 411
+ S F K G + A N D T L++N++N +G T I
Sbjct: 149 FNEDGSFRQSVFFQKTGTAYIATALRAARNADPNTKLYINDFNI-----EGTGAKSTGMI 203
Query: 412 EKIKEIQSVNSQLPLGIGLESH-----FPNSPPNLPYMRASLDTLRATGLPIWITELDV- 465
++ +Q N + GIG+++H P+S ++ +L G+ + ITELD+
Sbjct: 204 NLVRSLQQQNVPID-GIGVQAHLIVGQIPSS------IQQNLQNFANLGVEVAITELDIR 256
Query: 466 -------ASQPNQALYFEQVLREAHSHPGIQGIVMWT-----AWSP---QGCYRICLTDN 510
Q + V+R + G+ +W +W P G C D
Sbjct: 257 MTLPVTQQKLEQQQEDYRTVIRACKAVSRCVGVTVWDWTDRYSWVPGVFNGEGAACPWDE 316
Query: 511 NFKNLPA 517
N PA
Sbjct: 317 NLAKKPA 323
>XYNA_CALSR (P40944) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 684
Score = 89.7 bits (221), Expect = 2e-17
Identities = 95/389 (24%), Positives = 161/389 (40%), Gaps = 65/389 (16%)
Query: 112 LRKGLHYSLSAWI-QVSEETVPVTAVVKTTKGFKFGGAIFAEPNC----WSMLKGGLIAD 166
L+KG Y S + Q S + V +A +K G + A+ N W+ L L D
Sbjct: 253 LQKGGTYDFSLLVYQDSSKPVNFSAGIKLNDGKSTKEIVLAKQNVAPKKWTQLFATLDLD 312
Query: 167 TTGVAE--LYFESNNTSVEIWVDNVSLQPFTEKQWRSHQELSIEKDRKRKVVVRAVNEQG 224
T A+ +F ++ ++D S+ +++ W G
Sbjct: 313 TRFSAKDVSFFVKPAAAISYYLDLYSI---SDENW------------------------G 345
Query: 225 HPLPNASI-SLTMKRPG-FPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYA 282
P+P+ ++ SL K F G A+ L N + F T ENEMK + +
Sbjct: 346 QPVPDYNLPSLCEKYKNYFKIGVAVPYRALTNPVDVEVIKRHFNSITPENEMKPESLQPY 405
Query: 283 QGKDNYFDADAMLGFAEKQGIAVRGHNIFWDDPQYQPNWV------------SSLSPDQL 330
+G ++ AD + F +K I++RGH + W Q P+W S + L
Sbjct: 406 EGGFSFSIADEYVDFCKKDNISLRGHTLVWH--QQTPSWFFTNPETGEKLTNSEKDKEIL 463
Query: 331 NDAVEKRVNSIVSRYKGQLIGWDVVNENL--------HFSFFESKLGQNFSARMFNEVHN 382
D ++K + ++V RYKG++ WDVVNE + S + + LG + + F H
Sbjct: 464 LDRLKKHIQTVVGRYKGKVYAWDVVNEAIDENQPDGYRRSDWYNILGPEYIEKAFIWAHE 523
Query: 383 IDGQTTLFMNEYNTIEDSRDGLSTPPTYIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLP 442
D + LF N+Y+T + + +I K+ + G+GL+ H P++
Sbjct: 524 ADPKAKLFYNDYSTEDPYK------REFIYKLIKNLKAKGVPVHGVGLQCHISLDWPDVS 577
Query: 443 YMRASLDTL-RATGLPIWITELDVASQPN 470
+ ++ R GL I TE+D++ N
Sbjct: 578 EIEETVKLFSRIPGLEIHFTEIDISIAKN 606
>GUX_CELFI (P07986) Exoglucanase/xylanase precursor [Includes:
Exoglucanase (EC 3.2.1.91) (Exocellobiohydrolase)
(1,4-beta-cellobiohydrolase) (Beta-1,4-glycanase CEX);
Endo-1,4-beta-xylanase B (EC 3.2.1.8) (Xylanase B)]
Length = 484
Score = 86.3 bits (212), Expect = 2e-16
Identities = 66/272 (24%), Positives = 114/272 (41%), Gaps = 27/272 (9%)
Query: 240 GFPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAE 299
G FG A++ N L+ Y+ S F + EN MKW E +Q ++ D + +A
Sbjct: 53 GRDFGFALDPNRLSEAQYKAIADSEFNLVVAENAMKWDATEPSQNSFSFGAGDRVASYAA 112
Query: 300 KQGIAVRGHNIFWDDPQYQPNWVSSLSPDQLNDAVEKRVNSIVSRYKGQLIGWDVVNENL 359
G + GH + W P+W +L+ A+ V + ++G++ WDVVNE
Sbjct: 113 DTGKELYGHTLVWH--SQLPDWAKNLNGSAFESAMVNHVTKVADHFEGKVASWDVVNEAF 170
Query: 360 -------HFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNTIEDSRDGLSTPPTYI- 411
S F+ KLG + F D L +N+YN +G++ +
Sbjct: 171 ADGDGPPQDSAFQQKLGNGYIETAFRAARAADPTAKLCINDYNV-----EGINAKSNSLY 225
Query: 412 EKIKEIQSVNSQLPLGIGLESH-FPNSPPNLPYMRASLDTLRATGLPIWITELDVASQ-- 468
+ +K+ ++ L +G +SH P R +L G+ + ITELD+ +
Sbjct: 226 DLVKDFKARGVPLDC-VGFQSHLIVGQVPG--DFRQNLQRFADLGVDVRITELDIRMRTP 282
Query: 469 ------PNQALYFEQVLREAHSHPGIQGIVMW 494
QA +++V++ QG+ +W
Sbjct: 283 SDATKLATQAADYKKVVQACMQVTRCQGVTVW 314
>XYNA_PRERU (P48789) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 369
Score = 84.7 bits (208), Expect = 6e-16
Identities = 65/241 (26%), Positives = 111/241 (45%), Gaps = 37/241 (15%)
Query: 255 NAYQDW---------FASRFTVTTFENEMKWYTNEYAQGKDNYFD---ADAMLGFAEKQG 302
N++Q W F EN MK E +++Y+D AD ++ FAE+
Sbjct: 41 NSHQIWTHDPKIVHAITDNFNSVVAENCMK---GEIIHPEEDYYDWHDADQLVKFAEQHK 97
Query: 303 IAVRGHNIFWDDPQYQPNWV------SSLSPDQLNDAVEKRVNSIVSRYKGQLIGWDVVN 356
+ V GH + W P W+ ++ + L D + + ++V RYKG++ GWDVVN
Sbjct: 98 MTVHGHCLVWHSQA--PKWMFTDKEGKEVTREVLIDRMYHHITNVVKRYKGKIKGWDVVN 155
Query: 357 ENL------HFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYN-TIEDSRDGLSTPPT 409
E + S + +G +F F H D L+ N+Y+ +I R+ +
Sbjct: 156 EAILDNGEYRQSPYYKIIGPDFIKLAFIFAHQADPDAELYYNDYSMSIPAKRNAV----- 210
Query: 410 YIEKIKEIQSVNSQLPLGIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITELDVASQP 469
++ +KE+++ ++ +G++SH + PNL S+ A G+ + TELDV P
Sbjct: 211 -VKLVKELKAAGCRID-AVGMQSHNGFNYPNLEDYENSIKAFIAAGVDVQFTELDVNMLP 268
Query: 470 N 470
N
Sbjct: 269 N 269
>XYN2_BACST (P45703) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase)
Length = 330
Score = 84.3 bits (207), Expect = 8e-16
Identities = 67/244 (27%), Positives = 117/244 (47%), Gaps = 33/244 (13%)
Query: 241 FPFGSAINKNILNNNAYQDWFASRFTVTTFENEMKWYTNEYAQGKDNYFDADAMLGFAEK 300
F G+A+N L A Q T EN MK+ E+ Q ++ F D + +
Sbjct: 16 FRIGAAVNPVTLE--AQQSLLIRHVNSLTAENHMKF---EHLQPEEGRFTFDIAIKSSTS 70
Query: 301 --QGIAVRGHNIFWDDPQYQPNWVSSLSP------DQLNDAVEKRVNSIVSRYKGQLIGW 352
VRGH + W + P+WV S D L + ++ ++++V RYKG++ W
Sbjct: 71 PFSSHGVRGHTLVWHNQT--PSWVFQDSQGHFVGRDVLLERMKSHISTVVQRYKGKVYCW 128
Query: 353 DVVNEN--------LHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNT-IEDSRDG 403
DV+NE L S + +G +F + F H D + LF N+YN + R+
Sbjct: 129 DVINEAVADEGSEWLRSSTWRQIIGDDFIQQAFLYAHEADPEALLFYNDYNECFPEKREK 188
Query: 404 LSTPPTYIEKIKEIQSVNSQLPL-GIGLESHFPNSPPNLPYMRASLDTLRATGLPIWITE 462
+ T +K ++ + +P+ GIG+++H+ + P L +RA+++ + G+ + ITE
Sbjct: 189 IYT------LVKSLR--DKGIPIHGIGMQAHWSLNRPTLDEIRAAIERYASLGVILHITE 240
Query: 463 LDVA 466
LD++
Sbjct: 241 LDIS 244
>GUNB_CALSA (P10474) Endoglucanase/exoglucanase B precursor
[Includes: Endoglucanase (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Cellulase)
(Cellobiohydrolase); Exoglucanase (EC 3.2.1.91)
(Exocellobiohydrolase) (1,4-beta-cellobiohydrolase)]
Length = 1039
Score = 84.0 bits (206), Expect = 1e-15
Identities = 72/247 (29%), Positives = 111/247 (44%), Gaps = 31/247 (12%)
Query: 241 FPFGSAINKNILNNNAYQDWFASRFTVTTFENEMK---WYTNEYAQGKDNYFD-ADAMLG 296
F G AI L+N+ + F T ENEMK + + G F ADA +
Sbjct: 53 FMIGVAIPARCLSNDTDKRMVLKHFNSITAENEMKPESLLAGQTSTGLSYRFSTADAFVD 112
Query: 297 FAEKQGIAVRGHNIFWDDPQYQPNWV------SSLSPDQLNDAVEKRVNSIVSRYKGQLI 350
FA I +RGH + W + P+W LS D L +++ + +V RYKG++
Sbjct: 113 FASTNKIGIRGHTLVWHNQT--PDWFFKDSNGQRLSKDALLARLKQYIYDVVGRYKGKVY 170
Query: 351 GWDVVNE--------NLHFSFFESKLGQNFSARMFNEVHNIDGQTTLFMNEYNT-IEDSR 401
WDVVNE + S + G + + F H D LF N+YNT I R
Sbjct: 171 AWDVVNEAIDENQPDSYRRSTWYEICGPEYIEKAFIWAHEADPNAKLFYNDYNTEISKKR 230
Query: 402 DGLSTPPTYIEKIKEIQSVNSQLPL-GIGLESHFPNSPPNLPYMRASLDTLRA-TGLPIW 459
D + +K ++S +P+ GIG++ H + P++ + S+ + G+ I
Sbjct: 231 DFI------YNMVKNLKS--KGIPIHGIGMQCHINVNWPSVSEIENSIKLFSSIPGIEIH 282
Query: 460 ITELDVA 466
ITELD++
Sbjct: 283 ITELDMS 289
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 73,228,567
Number of Sequences: 164201
Number of extensions: 3252948
Number of successful extensions: 7187
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 7072
Number of HSP's gapped (non-prelim): 50
length of query: 588
length of database: 59,974,054
effective HSP length: 116
effective length of query: 472
effective length of database: 40,926,738
effective search space: 19317420336
effective search space used: 19317420336
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146757.5


