

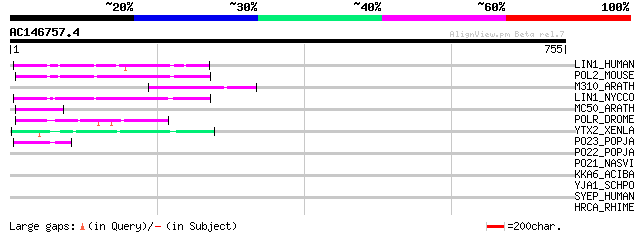
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.4 - phase: 0 /pseudo
(755 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 85 8e-16
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 84 2e-15
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 82 4e-15
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 77 1e-13
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 58 8e-08
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 57 2e-07
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 55 7e-07
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 48 8e-05
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 41 0.010
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 41 0.013
KKA6_ACIBA (P09885) Aminoglycoside 3'-phosphotransferase (EC 2.7... 34 1.2
YJA1_SCHPO (Q9USN4) Putative transporter C1529.01 32 6.2
SYEP_HUMAN (P07814) Bifunctional aminoacyl-tRNA synthetase [Incl... 32 8.1
HRCA_RHIME (Q92SK1) Heat-inducible transcription repressor hrcA 32 8.1
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 84.7 bits (208), Expect = 8e-16
Identities = 78/274 (28%), Positives = 124/274 (44%), Gaps = 20/274 (7%)
Query: 6 IMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQI 65
I+ NG+K + G RQG PLSP L I ++ L+ I + E K ++ G+ ++
Sbjct: 640 IILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIRQEKEIK---GIQLGKEEVKL 696
Query: 66 SHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLREDIL 125
S LFADD++++ E I A +L + F + SG KIN K+ + N + Q I+
Sbjct: 697 S--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTN-NRQTESQIM 753
Query: 126 HHTGFNQVNSLGMYLGANLAPGRSLRGKFNH----IVNKIQSKLNGWKQQCLTLAGRITL 181
F + YLG L R ++ F ++N+I+ N WK + GRI +
Sbjct: 754 SELPFTIASKRIKYLGIQLT--RDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINI 811
Query: 182 AKSVI--STIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKAD 239
K I I ++ K+P T E++K FIW ++AH+ + KA
Sbjct: 812 VKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQ----KRAHIAKSTLSQKNKA- 866
Query: 240 GGLGFRQTHKMNEAFLMKILWNLIKNPE-DLWCR 272
GG+ +A + K W +N + D W R
Sbjct: 867 GGITLPDFKLYYKATVTKTAWYWYQNRDIDQWNR 900
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 83.6 bits (205), Expect = 2e-15
Identities = 76/271 (28%), Positives = 122/271 (44%), Gaps = 18/271 (6%)
Query: 9 NGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISHL 68
NGEK + G RQG PLSPYLF I ++ L+ I Q E K ++ G+ +IS
Sbjct: 670 NGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQKEIK---GIQIGKEEVKIS-- 724
Query: 69 LFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVY-FSKNVDTQLREDILHH 127
L ADD++++ +L+ ++ F + G KIN NK+ + ++KN Q ++I
Sbjct: 725 LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKN--KQAEKEIRET 782
Query: 128 TGFNQVNSLGMYLGANLAPG-RSLRGK-FNHIVNKIQSKLNGWKQQCLTLAGRITLAKSV 185
T F+ V + YLG L + L K F + +I+ L WK + GRI + K
Sbjct: 783 TPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMA 842
Query: 186 ISTIPYYHMQY--AKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGGLG 243
I Y KIP +E++ F+W + K I+ + + GG+
Sbjct: 843 ILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNN-----KKPRIAKSLLKDKRTSGGIT 897
Query: 244 FRQTHKMNEAFLMKILWNLIKNPE-DLWCRV 273
A ++K W ++ + D W R+
Sbjct: 898 MPDLKLYYRAIVIKTAWYWYRDRQVDQWNRI 928
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 82.4 bits (202), Expect = 4e-15
Identities = 45/150 (30%), Positives = 75/150 (50%), Gaps = 4/150 (2%)
Query: 189 IPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKAD-GGLGFRQT 247
+P Y M ++ K +C ++ F W + RK ++W C K D GGLGFR
Sbjct: 3 LPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRDL 62
Query: 248 HKMNEAFLMKILWNLIKNPEDLWCRVLRSKYGRNNDLI-ASINAHPYDSPLWKALVNIWN 306
N+A L K + +I P L R+LRS+Y ++ ++ S+ P S W+++++
Sbjct: 63 GWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRP--SYAWRSIIHGRE 120
Query: 307 DFKGHVVWNIGDGRNTNFWLDKWVPNNESL 336
++ IGDG +T WLD+W+ + L
Sbjct: 121 LLSRGLLRTIGDGIHTKVWLDRWIMDETPL 150
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 77.4 bits (189), Expect = 1e-13
Identities = 74/273 (27%), Positives = 118/273 (43%), Gaps = 16/273 (5%)
Query: 6 IMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQI 65
I+ NG K +F G RQG PLSP LF I M+ L+ I ++ K + G +I
Sbjct: 640 IILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREEKAIK---GIHIG--SEEI 694
Query: 66 SHLLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQLREDIL 125
LFADD++++ E + + +L + + SG KIN +K+ + N + Q + +
Sbjct: 695 KLSLFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTN-NNQAEKTVK 753
Query: 126 HHTGFNQVNSLGMYLGANLAPGRSLRGKFNH--IVNKIQSKLNGWKQQCLTLAGRITLAK 183
F V YLG L K N+ + +I +N WK + GRI + K
Sbjct: 754 DSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVK 813
Query: 184 SVISTIPYYHMQY--AKIPKTICDEIDKIQRGFIWGDSDQGRKAHLISWDVCCLPKADGG 241
I Y+ K P + +++KI FIW +K I+ + GG
Sbjct: 814 MSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIW-----NQKKPQIAKTLLSNKNKAGG 868
Query: 242 LGFRQTHKMNEAFLMKILWNLIKNPE-DLWCRV 273
+ ++ ++K W KN E D+W R+
Sbjct: 869 ITLPDLRLYYKSIVIKTAWYWHKNREVDVWNRI 901
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 58.2 bits (139), Expect = 8e-08
Identities = 29/65 (44%), Positives = 38/65 (57%)
Query: 9 NGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISHL 68
NG +RG+RQGDPLSPYLF++C + LS + E +R P+I+HL
Sbjct: 15 NGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHL 74
Query: 69 LFADD 73
LFADD
Sbjct: 75 LFADD 79
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 56.6 bits (135), Expect = 2e-07
Identities = 58/216 (26%), Positives = 94/216 (42%), Gaps = 22/216 (10%)
Query: 9 NGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISH- 67
+G ++ F RG++QGDPLSP LF + MDRL + ++ AK G I++
Sbjct: 526 DGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIGAK---------VGNAITNA 576
Query: 68 LLFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKTCVYFSKNVDTQ---LREDI 124
FADDL+LFAE + + LD F G K+N +K K Q + E
Sbjct: 577 AAFADDLVLFAETRMGLQVLLDKTLD-FLSIVGLKLNADKCFTVGIKGQPKQKCTVLEAQ 635
Query: 125 LHHTGFNQVNSL-----GMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWKQQCLTLAGRI 179
+ G +++ SL YLG N +R + I KL + L R+
Sbjct: 636 SFYVGSSEIPSLKRTDEWKYLGINFTATGRVR---CNPAEDIGPKLQRLTKAPLKPQQRL 692
Query: 180 TLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFI 215
++V+ Y+ + + + + DK+ R ++
Sbjct: 693 FALRTVLIPQLYHKLALGSVAIGVLRKTDKLIRYYV 728
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 55.1 bits (131), Expect = 7e-07
Identities = 61/291 (20%), Positives = 116/291 (38%), Gaps = 40/291 (13%)
Query: 3 EYIIMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMD--------RLSHMIADQVEAKYWI 54
E ++ N T RG+RQG PLS L+ + ++ RL+ ++ + + + +
Sbjct: 632 ECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKRLTGLVLKEPDMRVVL 691
Query: 55 PMRAGRYGPQISHLLFADDLLLFAE--ASIEQAHCVLHCLDIFCQASGQKINRNKTCVYF 112
+ADD++L A+ +E+A C +++ AS +IN +K+
Sbjct: 692 S-------------AYADDVILVAQDLVDLERAQ---ECQEVYAAASSARINWSKSSGLL 735
Query: 113 SKNVDTQLREDILHHTGFNQ--VNSLGMYLGANLAPGRSLRGKFNHIVNKIQSKLNGWK- 169
++ + + LG+YL A P + F + + ++L WK
Sbjct: 736 EGSLKVDFLPPAFRDISWESKIIKYLGVYLSAEEYP---VSQNFIELEECVLTRLGKWKG 792
Query: 170 -QQCLTLAGRITLAKSVISTIPYYHMQYAKIPKTICDEIDKIQRGFIWGDSDQGRKAHLI 228
+ L++ GR + ++++ +Y + + +I + F+W H +
Sbjct: 793 FAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGK------HWV 846
Query: 229 SWDVCCLPKADGGLGFRQTHKMNEAF-LMKILWNLIKNPEDLWCRVLRSKY 278
S V LP +GG G F L +I L +P WC + S Y
Sbjct: 847 SAGVSSLPLKEGGQGVVCIRSQVHTFRLQQIQRYLYADPSPQWCTLASSFY 897
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 48.1 bits (113), Expect = 8e-05
Identities = 28/78 (35%), Positives = 40/78 (50%), Gaps = 6/78 (7%)
Query: 6 IMWNGEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQI 65
I+ G T+ + G++QGDPLSP LF I +D L + D+ P +I
Sbjct: 70 IVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASMTP------ACKI 123
Query: 66 SHLLFADDLLLFAEASIE 83
+ L FADDLLL + I+
Sbjct: 124 ASLAFADDLLLLEDRDID 141
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 41.2 bits (95), Expect = 0.010
Identities = 32/100 (32%), Positives = 45/100 (45%), Gaps = 9/100 (9%)
Query: 10 GEKTDTFFSTRGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQ-ISHL 68
G +T RG++QGDPLSP+LF +D L + P G G + I L
Sbjct: 189 GSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQS-------TPGIGGTIGEEKIPVL 241
Query: 69 LFADDLLLFAEASIEQAHCVLHCLDIFCQASGQKINRNKT 108
FADDLLL + + L + F + G +N K+
Sbjct: 242 AFADDLLLLEDNDV-LLPTTLATVANFFRLRGMSLNAKKS 280
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 40.8 bits (94), Expect = 0.013
Identities = 26/60 (43%), Positives = 33/60 (54%), Gaps = 8/60 (13%)
Query: 20 RGIRQGDPLSPYLFVICMDRLSHMIADQVEAKYWIPMRAGRYGPQISHLLFADDLLLFAE 79
RG+RQGDPLSP LF MD + + + M A + G L+FADDL+L AE
Sbjct: 511 RGVRQGDPLSPLLFNCVMDAVLRRLPENT----GFLMGAEKIGA----LVFADDLVLLAE 562
>KKA6_ACIBA (P09885) Aminoglycoside 3'-phosphotransferase (EC
2.7.1.95) (Kanamycin kinase, type VI)
(Neomycin-kanamycin phosphotransferase, type VI)
(APH(3')VI)
Length = 259
Score = 34.3 bits (77), Expect = 1.2
Identities = 18/66 (27%), Positives = 38/66 (57%), Gaps = 8/66 (12%)
Query: 272 RVLRSKYGRNNDLIASINAHPYDSPLW---KALVNIWNDF-----KGHVVWNIGDGRNTN 323
R+ SK+ +N L+ I+ +D+ LW K +++WN+ + +V++ GD ++N
Sbjct: 133 RLKESKFFIDNQLLDDIDQDDFDTELWGDHKTYLSLWNELTETRVEERLVFSHGDITDSN 192
Query: 324 FWLDKW 329
++DK+
Sbjct: 193 IFIDKF 198
>YJA1_SCHPO (Q9USN4) Putative transporter C1529.01
Length = 462
Score = 32.0 bits (71), Expect = 6.2
Identities = 30/133 (22%), Positives = 59/133 (43%), Gaps = 22/133 (16%)
Query: 589 SGLEKVGSSLIVMVLARIWGLLLDVAAFFVIQTVDGSKATLRRLELVM--PYMLRCGDYI 646
+GL + S + ++ ++ G+ +D ++ + +G A R+ +++ + G +I
Sbjct: 315 AGLNYIASGIGLIFGSQASGIFIDKTFRYLKRRNNGKMAPEFRVPVILLGTFFFPAGLFI 374
Query: 647 WVWT------WLGG-------NITLIL*WKVIQRF*ST*FLIISSSMGIFLF*FTVSGIC 693
+ WT W+G NI L+L W+ IQ + F+I ++S T C
Sbjct: 375 YGWTAQYHTHWIGPDIGAAMFNIGLMLGWRGIQTYLIDSFMIYAASS-------TAVACC 427
Query: 694 *R*VGMFNSPILG 706
R + F P+ G
Sbjct: 428 VRSIAAFAFPLFG 440
>SYEP_HUMAN (P07814) Bifunctional aminoacyl-tRNA synthetase [Includes:
Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA
ligase); Prolyl-tRNA synthetase (EC 6.1.1.15)
(Proline--tRNA ligase)]
Length = 1440
Score = 31.6 bits (70), Expect = 8.1
Identities = 17/62 (27%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Query: 413 HQDCPILEGDWKSIWKWHGPH-----RIQTFIWLDTHERILTNFQRSNGVVEYL-LIARI 466
H+D PI W ++ +W H R + F+W + H T + + V++ L L A++
Sbjct: 1065 HRDLPIKLNQWCNVVRWEFKHPQPFLRTREFLWQEGHSAFATMEEAAEEVLQILDLYAQV 1124
Query: 467 VE 468
E
Sbjct: 1125 YE 1126
>HRCA_RHIME (Q92SK1) Heat-inducible transcription repressor hrcA
Length = 359
Score = 31.6 bits (70), Expect = 8.1
Identities = 29/95 (30%), Positives = 42/95 (43%), Gaps = 10/95 (10%)
Query: 144 LAPGRSLR---GKFNHIVNKIQSKLNGWKQQCLTLAGRITLAKSVISTIPYYHMQYAKIP 200
LAP ++L G+ + + N+I G LT A A TIP Q K+
Sbjct: 156 LAPTKALAVLVGEHDQVENRIIELPAGITSAQLTEAANFVNAHLAGQTIPELRTQLEKVK 215
Query: 201 KTICDEIDK-----IQRGF-IW-GDSDQGRKAHLI 228
+T+ E+D ++RG IW G G+ A LI
Sbjct: 216 ETVRGELDALSQDLVERGLAIWSGSEGDGQPARLI 250
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.338 0.149 0.503
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 85,274,192
Number of Sequences: 164201
Number of extensions: 3559471
Number of successful extensions: 10658
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 10634
Number of HSP's gapped (non-prelim): 17
length of query: 755
length of database: 59,974,054
effective HSP length: 118
effective length of query: 637
effective length of database: 40,598,336
effective search space: 25861140032
effective search space used: 25861140032
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146757.4


