

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146757.11 - phase: 0 /pseudo
(348 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
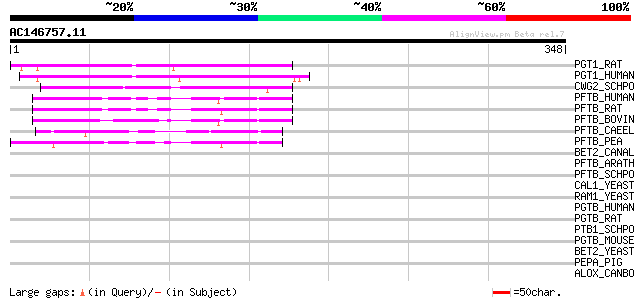
Score E
Sequences producing significant alignments: (bits) Value
PGT1_RAT (P53610) Geranylgeranyl transferase type I beta subunit... 116 7e-26
PGT1_HUMAN (P53609) Geranylgeranyl transferase type I beta subun... 113 6e-25
CWG2_SCHPO (P32434) Type I protein geranylgeranyltransferase bet... 58 3e-08
PFTB_HUMAN (P49356) Protein farnesyltransferase beta subunit (EC... 58 4e-08
PFTB_RAT (Q02293) Protein farnesyltransferase beta subunit (EC 2... 57 5e-08
PFTB_BOVIN (P49355) Protein farnesyltransferase beta subunit (EC... 55 2e-07
PFTB_CAEEL (P41992) Probable protein farnesyltransferase beta su... 55 3e-07
PFTB_PEA (Q04903) Protein farnesyltransferase beta subunit (EC 2... 45 3e-04
BET2_CANAL (O93830) Type II proteins geranylgeranyltransferase b... 43 0.001
PFTB_ARATH (Q38920) Protein farnesyltransferase beta subunit (EC... 42 0.002
PFTB_SCHPO (O13782) Protein farnesyltransferase beta subunit (EC... 42 0.002
CAL1_YEAST (P18898) Type I proteins geranylgeranyltransferase be... 42 0.002
RAM1_YEAST (P22007) Protein farnesyltransferase beta subunit (EC... 40 0.007
PGTB_HUMAN (P53611) Geranylgeranyl transferase type II beta subu... 38 0.033
PGTB_RAT (Q08603) Geranylgeranyl transferase type II beta subuni... 36 0.13
PTB1_SCHPO (P46960) Type II proteins geranylgeranyltransferase b... 35 0.28
PGTB_MOUSE (P53612) Geranylgeranyl transferase type II beta subu... 35 0.28
BET2_YEAST (P20133) Type II proteins geranylgeranyltransferase b... 34 0.62
PEPA_PIG (P00791) Pepsin A precursor (EC 3.4.23.1) 32 2.4
ALOX_CANBO (Q00922) Alcohol oxidase (EC 1.1.3.13) (AOX) (Methano... 32 3.1
>PGT1_RAT (P53610) Geranylgeranyl transferase type I beta subunit
(EC 2.5.1.59) (Type I protein geranyl-geranyltransferase
beta subunit) (GGTase-I-beta)
Length = 377
Score = 116 bits (291), Expect = 7e-26
Identities = 74/186 (39%), Positives = 106/186 (56%), Gaps = 11/186 (5%)
Query: 1 MSSSEE---ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDI 55
M+++E+ A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+
Sbjct: 1 MAATEDDRLAGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDM 60
Query: 56 LNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFP--PDEN-GVFH-HN 111
L+SL +V K+ + W+ S QV T D +N GF GS P P +N G H ++
Sbjct: 61 LDSLDVVNKDDIIEWIYSLQVL--PTEDRSNLDRCGFRGSSYLGIPFNPSKNPGTAHPYD 118
Query: 112 NSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+ H+A TY L+ L I+G DLS +D E+ + ++ LQ DGSF + G E D+RFVYCA
Sbjct: 119 SGHIAMTYTGLSCLIILGDDLSRVDKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCA 178
Query: 172 GWITGM 177
I M
Sbjct: 179 SCICYM 184
>PGT1_HUMAN (P53609) Geranylgeranyl transferase type I beta subunit
(EC 2.5.1.59) (Type I protein geranyl-geranyltransferase
beta subunit) (GGTase-I-beta)
Length = 377
Score = 113 bits (283), Expect = 6e-25
Identities = 74/192 (38%), Positives = 105/192 (54%), Gaps = 12/192 (6%)
Query: 7 ASSMETTTID--KDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEK 64
A S E +D +D HV F Q +LP Y S E + LT+A+F +S LD+L+SL +V K
Sbjct: 10 AGSGEGERLDFLRDRHVRFFQRCLQVLPERYSSLETSRLTIAFFALSGLDMLDSLDVVNK 69
Query: 65 EAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDEN---GVFH-HNNSHLASTYC 120
+ + W+ S QV T D +N GF GS P + + G H +++ H+A TY
Sbjct: 70 DDIIEWIYSLQVL--PTEDRSNLNRCGFRGSSYLGIPFNPSKAPGTAHPYDSGHIAMTYT 127
Query: 121 ALAILKIVGYDLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM--E 178
L+ L I+G DLS ++ E+ + ++ LQ DGSF + G E D+RFVYCA I M
Sbjct: 128 GLSCLVILGDDLSRVNKEACLAGLRALQLEDGSFCAVPEGSENDMRFVYCASCICYMLNN 187
Query: 179 WT--RKKSRITY 188
W+ K ITY
Sbjct: 188 WSGMDMKKAITY 199
>CWG2_SCHPO (P32434) Type I protein geranylgeranyltransferase beta
subunit (EC 2.5.1.59) (Type I protein
geranyl-geranyltransferase beta subunit) (GGTase-I-beta)
(PGGT)
Length = 355
Score = 58.2 bits (139), Expect = 3e-08
Identities = 37/160 (23%), Positives = 73/160 (45%), Gaps = 8/160 (5%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRG 79
H+ F + L P PYE + LA+F + LD+LN+L+ ++ + +W+ + +
Sbjct: 8 HIAFFKRHLILFPTPYEEHDCERTVLAFFCLLGLDLLNALNTIDDDDKKSWI-EWIYKNY 66
Query: 80 TTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSES 139
T + ++ GF +T P + LA T ++ L +G +LS +D +
Sbjct: 67 VTKESKGIKYSGFQAYRTGIQP-----ISFEQEPQLAGTVFSICCLLFLGDNLSRIDRDL 121
Query: 140 MSSSMKNLQQPDGSFMPIHIG--GETDLRFVYCAGWITGM 177
+ + ++ + G F I + + D+R +Y A I +
Sbjct: 122 IKNFVELCKTSQGHFRSIAVPSCSDQDMRQLYMATTIASL 161
>PFTB_HUMAN (P49356) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta)
Length = 437
Score = 57.8 bits (138), Expect = 4e-08
Identities = 46/167 (27%), Positives = 77/167 (45%), Gaps = 30/167 (17%)
Query: 15 IDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSF 74
+ ++ H + + L + YE + + L Y+++ SL++L+ + VA V F
Sbjct: 73 LQREKHFHYLKRGLRQLTDAYECLDASRPWLCYWILHSLELLDE---PIPQIVATDVCQF 129
Query: 75 QVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVG----Y 130
P G G Q+P HLA TY A+ L I+G Y
Sbjct: 130 LE---LCQSPEGG-----FGGGPGQYP------------HLAPTYAAVNALCIIGTEEAY 169
Query: 131 DLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
D+ ++ E + + +L+QPDGSF+ +H+GGE D+R YCA + +
Sbjct: 170 DI--INREKLLQYLYSLKQPDGSFL-MHVGGEVDVRSAYCAASVASL 213
>PFTB_RAT (Q02293) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta)
Length = 437
Score = 57.4 bits (137), Expect = 5e-08
Identities = 44/165 (26%), Positives = 77/165 (46%), Gaps = 26/165 (15%)
Query: 15 IDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSF 74
+ ++ H + + L + YE + + L Y+++ SL++L+ + VA V F
Sbjct: 73 LQREKHFHYLKRGLRQLTDAYECLDASRPWLCYWILHSLELLDE---PIPQIVATDVCQF 129
Query: 75 QVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYD--L 132
P+ G G Q+P HLA TY A+ L I+G +
Sbjct: 130 LE---LCQSPDGG-----FGGGPGQYP------------HLAPTYAAVNALCIIGTEEAY 169
Query: 133 SSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
+ ++ E + + +L+QPDGSF+ +H+GGE D+R YCA + +
Sbjct: 170 NVINREKLLQYLYSLKQPDGSFL-MHVGGEVDVRSAYCAASVASL 213
>PFTB_BOVIN (P49355) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta)
Length = 437
Score = 55.5 bits (132), Expect = 2e-07
Identities = 46/167 (27%), Positives = 77/167 (45%), Gaps = 30/167 (17%)
Query: 15 IDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSF 74
+ ++ H + + L + YE + + L Y+++ SL++L+ E + V +
Sbjct: 73 LQREKHFHYLKRGLRQLTDAYECLDASRPWLCYWILHSLELLD-------EPIPQMVATD 125
Query: 75 QVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVG----Y 130
Q G F G G Q+P HLA TY A+ L I+G Y
Sbjct: 126 VCQFLELCQSPEGGFGGGPG----QYP------------HLAPTYAAVNALCIIGTEEAY 169
Query: 131 DLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
D+ ++ E + + +L+QPDGSF+ +H GGE D+R YCA + +
Sbjct: 170 DV--INREKLLQYLYSLKQPDGSFL-MHDGGEVDVRSAYCAASVASL 213
>PFTB_CAEEL (P41992) Probable protein farnesyltransferase beta
subunit (EC 2.5.1.58) (CAAX farnesyltransferase beta
subunit) (RAS proteins prenyltransferase beta)
(FTase-beta)
Length = 335
Score = 54.7 bits (130), Expect = 3e-07
Identities = 44/157 (28%), Positives = 68/157 (43%), Gaps = 29/157 (18%)
Query: 17 KDLHVTFAQMMYHLLPNPYESQEINHLTLA--YFVISSLDILNSLHLVEKEAVANWVLSF 74
KDLH F Y N Y HL ++ Y+ ++++D+ L + E + N+VL
Sbjct: 26 KDLHANFINQ-YEKNKNSYHYIMAEHLRVSGIYWCVNAMDLSKQLERMSTEEIVNYVLGC 84
Query: 75 QVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSS 134
+ D G G ++SHL T CA+ L I+ +
Sbjct: 85 R-----NTDGGYGPAPG-------------------HDSHLLHTLCAVQTL-IIFNSIEK 119
Query: 135 LDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
D++++S +K LQQ DGSF + GE D RF C+
Sbjct: 120 ADADTISEYVKGLQQEDGSFCG-DLSGEVDTRFTLCS 155
>PFTB_PEA (Q04903) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta)
Length = 419
Score = 45.1 bits (105), Expect = 3e-04
Identities = 48/176 (27%), Positives = 77/176 (43%), Gaps = 29/176 (16%)
Query: 1 MSSSEEASSMETTTIDKDLHVTFAQM--MYHLLPN-PYESQEINHLTLAYFVISSLDILN 57
M +S A + T +D + +Q+ +Y L N P +Q I L Y++I S+ +L
Sbjct: 1 MEASTAAETPTPTVSQRDQWIVESQVFHIYQLFANIPPNAQSIIRPWLCYWIIHSIALLG 60
Query: 58 SLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLAS 117
++ + N V DPN G G Q P HLA+
Sbjct: 61 ES--IDDDLEDNTVDFLN----RCQDPNGGYAGG-----PGQMP------------HLAT 97
Query: 118 TYCALAILKIVGYD--LSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
TY A+ L +G + L+S++ + M+ ++QP+G F +H GE D+R Y A
Sbjct: 98 TYAAVNTLITLGGEKSLASINRNKLYGFMRRMKQPNGGFR-MHDEGEIDVRACYTA 152
>BET2_CANAL (O93830) Type II proteins geranylgeranyltransferase beta
subunit (EC 2.5.1.60) (Type II protein
geranyl-geranyltransferase beta subunit)
(GGTase-II-beta) (PGGT) (YPT1/SEC4 proteins
geranylgeranyltransferase beta subunit)
Length = 341
Score = 43.1 bits (100), Expect = 0.001
Identities = 29/76 (38%), Positives = 39/76 (51%), Gaps = 11/76 (14%)
Query: 106 GVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSS----------MKNLQQPDGSFM 155
G F ++ H+ ST AL +LKI +L+ L+ + SS+ + LQ PDGSF
Sbjct: 81 GSFPKHDGHILSTLSALQVLKIYDQELTVLNDNNESSNGNKRERLIKFITGLQLPDGSFQ 140
Query: 156 PIHIGGETDLRFVYCA 171
G E D RFVY A
Sbjct: 141 GDKYG-EVDTRFVYTA 155
>PFTB_ARATH (Q38920) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta) (Enhanced
response to abscisic acid 1)
Length = 404
Score = 42.4 bits (98), Expect = 0.002
Identities = 41/159 (25%), Positives = 68/159 (41%), Gaps = 26/159 (16%)
Query: 15 IDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSF 74
I +D + + L + S + N L Y+++ S+ +L V+ E +N +
Sbjct: 3 IQRDKQLDYLMKGLRQLGPQFSSLDANRPWLCYWILHSIALLGET--VDDELESNAIDFL 60
Query: 75 QVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYD--L 132
+G+ G +G Q P HLA+TY A+ L +G D L
Sbjct: 61 GRCQGSE---------GGYGGGPGQLP------------HLATTYAAVNALVTLGGDKAL 99
Query: 133 SSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
SS++ E MS ++ ++ G F +H GE D+R Y A
Sbjct: 100 SSINREKMSCFLRRMKDTSGGFR-MHDMGEMDVRACYTA 137
>PFTB_SCHPO (O13782) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase) (FTase-beta)
Length = 382
Score = 42.0 bits (97), Expect = 0.002
Identities = 42/169 (24%), Positives = 75/169 (43%), Gaps = 27/169 (15%)
Query: 11 ETTTIDKDLHVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANW 70
E+ + + H+ + M LP+P+ + + + Y+ +SSL IL L +
Sbjct: 25 ESQSFNLQKHLKYLTKMLDPLPSPFTVLDASRAWMVYWELSSLAILGKL---DSSVCERA 81
Query: 71 VLSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCA-LAILKIVG 129
+ S + +G +G F G +G + HL STY + L+I
Sbjct: 82 ISSVRQLKGP-----SGGFCGGNG----------------QDEHLLSTYASILSICLCDS 120
Query: 130 YDLSSL-DSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCAGWITGM 177
D SL + + + + +L+ PDGSF ++ GE+D R VY A ++ +
Sbjct: 121 TDAYSLIERDRLYDWLFSLKNPDGSFR-VNNEGESDARSVYAAVCVSSL 168
>CAL1_YEAST (P18898) Type I proteins geranylgeranyltransferase beta
subunit (EC 2.5.1.59) (Type I protein
geranyl-geranyltransferase beta subunit) (GGTase-I-beta)
(PGGT) (RAS proteins geranylgeranyltransferase beta
subunit)
Length = 376
Score = 42.0 bits (97), Expect = 0.002
Identities = 41/164 (25%), Positives = 68/164 (41%), Gaps = 20/164 (12%)
Query: 20 HVTFAQMMYHLLPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRG 79
H F + LLP+ ++ ++N + + ++ IS L I + + W+ + +
Sbjct: 16 HRKFFERHLQLLPSSHQGHDVNRMAIIFYSISGLSIFDVNVSAKYGDHLGWMRKHYI-KT 74
Query: 80 TTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCAL---AILKIVGYDLSSLD 136
+D N GF GS P H +L +T AL +L+ Y + LD
Sbjct: 75 VLDDTENTVISGFVGSLVMNIP-------HATTINLPNTLFALLSMIMLRDYEYFETILD 127
Query: 137 SESMSSSMKNLQQPD-GSFM-----PIHIGGET---DLRFVYCA 171
S++ + Q+PD GSF+ + G DLRF Y A
Sbjct: 128 KRSLARFVSKCQRPDRGSFVSCLDYKTNCGSSVDSDDLRFCYIA 171
>RAM1_YEAST (P22007) Protein farnesyltransferase beta subunit (EC
2.5.1.58) (CAAX farnesyltransferase beta subunit) (RAS
proteins prenyltransferase beta) (FTase-beta)
Length = 431
Score = 40.4 bits (93), Expect = 0.007
Identities = 37/161 (22%), Positives = 70/161 (42%), Gaps = 26/161 (16%)
Query: 15 IDKDLHVTFAQMMYHL-LPNPYESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLS 73
+ K+ H + + + + LP + + + + Y+ I NSL +++++ +++
Sbjct: 78 LTKEFHKMYLDVAFEISLPPQMTALDASQPWMLYW------IANSLKVMDRDWLSDDTKR 131
Query: 74 FQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKI---VGY 130
V + T P+ G F G G SHLASTY A+ L + +
Sbjct: 132 KIVDKLFTISPSGGPFGGGPGQL----------------SHLASTYAAINALSLCDNIDG 175
Query: 131 DLSSLDSESMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
+D + + + +L++P+G F GE D R +YCA
Sbjct: 176 CWDRIDRKGIYQWLISLKEPNGGFKTCLEVGEVDTRGIYCA 216
>PGTB_HUMAN (P53611) Geranylgeranyl transferase type II beta subunit
(EC 2.5.1.60) (Rab geranylgeranyltransferase beta
subunit) (Rab geranyl-geranyltransferase beta subunit)
(Rab GG transferase beta) (Rab GGTase beta)
Length = 331
Score = 38.1 bits (87), Expect = 0.033
Identities = 37/138 (26%), Positives = 62/138 (44%), Gaps = 28/138 (20%)
Query: 35 YESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHG 94
Y E ++ Y+ ++ +D++ LH + +E + ++ S Q + G G
Sbjct: 39 YCMSEYLRMSGIYWGLTVMDLMGQLHRMNREEILAFIKSCQHECG-----------GISA 87
Query: 95 SKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYD-LSSLDSESMSSSMKNLQQPDGS 153
S + H + HL T A+ IL + YD ++ +D + +K LQ+ DGS
Sbjct: 88 S-----------IGH--DPHLLYTLSAVQILTL--YDSINVIDVNKVVEYVKGLQKEDGS 132
Query: 154 FMPIHIGGETDLRFVYCA 171
F I GE D RF +CA
Sbjct: 133 FAG-DIWGEIDTRFSFCA 149
>PGTB_RAT (Q08603) Geranylgeranyl transferase type II beta subunit
(EC 2.5.1.60) (Rab geranylgeranyltransferase beta
subunit) (Rab geranyl-geranyltransferase beta subunit)
(Rab GG transferase beta) (Rab GGTase beta)
Length = 331
Score = 36.2 bits (82), Expect = 0.13
Identities = 33/138 (23%), Positives = 62/138 (44%), Gaps = 28/138 (20%)
Query: 35 YESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHG 94
Y E ++ Y+ ++ +D++ LH + KE + ++ S Q + G +
Sbjct: 39 YCMSEYLRMSGVYWGLTVMDLMGQLHRMNKEEILVFIKSCQHECGGVSASIG-------- 90
Query: 95 SKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYD-LSSLDSESMSSSMKNLQQPDGS 153
++ HL T A+ IL + YD + ++ + + + +++LQ+ DGS
Sbjct: 91 ----------------HDPHLLYTLSAVQILTL--YDSIHVINVDKVVAYVQSLQKEDGS 132
Query: 154 FMPIHIGGETDLRFVYCA 171
F I GE D RF +CA
Sbjct: 133 FAG-DIWGEIDTRFSFCA 149
>PTB1_SCHPO (P46960) Type II proteins geranylgeranyltransferase beta
subunit (EC 2.5.1.60) (Type II protein
geranyl-geranyltransferase beta subunit)
(GGTase-II-beta)
Length = 311
Score = 35.0 bits (79), Expect = 0.28
Identities = 14/48 (29%), Positives = 30/48 (62%)
Query: 41 NHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQ 88
+H+T + + L +L+SLH+V+K+ VA++++ Q + G+ G+
Sbjct: 79 DHITNTVYAVQVLAMLDSLHVVDKDKVASYIIGLQNEDGSMKGDRWGE 126
Score = 31.2 bits (69), Expect = 4.0
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 41 NHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTN 82
+H + + +++L ILN L L+++E + W+ QV+ G N
Sbjct: 175 SHGAMVFTCVAALKILNKLDLIDEELLGWWISERQVKGGGLN 216
>PGTB_MOUSE (P53612) Geranylgeranyl transferase type II beta subunit
(EC 2.5.1.60) (Rab geranylgeranyltransferase beta
subunit) (Rab geranyl-geranyltransferase beta subunit)
(Rab GG transferase beta) (Rab GGTase beta)
Length = 339
Score = 35.0 bits (79), Expect = 0.28
Identities = 35/138 (25%), Positives = 64/138 (46%), Gaps = 28/138 (20%)
Query: 35 YESQEINHLTLAYFVISSLDILNSLHLVEKEAVANWVLSFQVQRGTTNDPNNGQFYGFHG 94
Y E ++ Y+ ++ +D++ LH + +E + ++ S Q + G G
Sbjct: 47 YCMSEYLRMSGVYWGLTVMDLMGQLHRMNREEILVFIKSCQHECG-----------GISA 95
Query: 95 SKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYD-LSSLDSESMSSSMKNLQQPDGS 153
S + H + HL T A+ IL + YD + ++ + + + +++LQ+ DGS
Sbjct: 96 S-----------IGH--DPHLLYTLSAVQILTL--YDSVHVINVDKVVAYVQSLQKEDGS 140
Query: 154 FMPIHIGGETDLRFVYCA 171
F I GE D RF +CA
Sbjct: 141 FAG-DIWGEIDTRFSFCA 157
>BET2_YEAST (P20133) Type II proteins geranylgeranyltransferase beta
subunit (EC 2.5.1.60) (Type II protein
geranyl-geranyltransferase beta subunit)
(GGTase-II-beta) (PGGT) (YPT1/SEC4 proteins
geranylgeranyltransferase beta subunit)
Length = 325
Score = 33.9 bits (76), Expect = 0.62
Identities = 44/162 (27%), Positives = 69/162 (42%), Gaps = 29/162 (17%)
Query: 14 TIDKDLHVTFAQMMYHLLPNPYESQEINHLTL--AYFVISSLDILNSLHLVEKEAVANWV 71
T+ K+ H+ + + + N +E HL L Y+ +++L +L+S KE V ++V
Sbjct: 6 TLLKEKHIRYIESLDTKKHN-FEYWLTEHLRLNGIYWGLTALCVLDSPETFVKEEVISFV 64
Query: 72 LSFQVQRGTTNDPNNGQFYGFHGSKTSQFPPDENGVFHHNNSHLASTYCALAILKIV-GY 130
LS K F P F +++HL +T A+ IL
Sbjct: 65 LSCW------------------DDKYGAFAP-----FPRHDAHLLTTLSAVQILATYDAL 101
Query: 131 DLSSLDSE-SMSSSMKNLQQPDGSFMPIHIGGETDLRFVYCA 171
D+ D + + S ++ Q DGSF G E D RFVY A
Sbjct: 102 DVLGKDRKVRLISFIRGNQLEDGSFQGDRFG-EVDTRFVYTA 142
>PEPA_PIG (P00791) Pepsin A precursor (EC 3.4.23.1)
Length = 386
Score = 32.0 bits (71), Expect = 2.4
Identities = 18/62 (29%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query: 95 SKTSQFPPDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQPD-GS 153
S +QF PD++ F + L+ TY ++ I+GYD + S ++ + L + + GS
Sbjct: 110 SDHNQFNPDDSSTFEATSQELSITYGTGSMTGILGYDTVQVGGISDTNQIFGLSETEPGS 169
Query: 154 FM 155
F+
Sbjct: 170 FL 171
>ALOX_CANBO (Q00922) Alcohol oxidase (EC 1.1.3.13) (AOX) (Methanol
oxidase) (MOX)
Length = 663
Score = 31.6 bits (70), Expect = 3.1
Identities = 37/168 (22%), Positives = 65/168 (38%), Gaps = 27/168 (16%)
Query: 6 EASSMETTTIDKDLHVTFAQMMYHLLPNPY---ESQEINHLTLAYFVISSLDILNSLHLV 62
E S+ +T + LH+T A + + P + + NH+T ++ + S DI +
Sbjct: 493 EQSAEDTKKVAGPLHLT-ANLYHGSWSTPIGEADKHDPNHVTSSHINVYSKDIQYTKE-- 549
Query: 63 EKEAVANWVL-------------SFQVQRGTTNDPNNG------QFYGFHGSKTSQFP-- 101
+ EA+ N++ S + G N P G +G G K +
Sbjct: 550 DDEAIENYIKEHAETTWHCLGTNSMAPREGNKNAPEGGVLDPRLNVHGVKGLKVADLSVC 609
Query: 102 PDENGVFHHNNSHLASTYCALAILKIVGYDLSSLDSESMSSSMKNLQQ 149
PD G + + A+ + + +GY S LD E +K +Q
Sbjct: 610 PDNVGCNTFSTALTIGEKAAVLVAEDLGYSGSELDMEVPQHKLKTYEQ 657
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.335 0.144 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,418,899
Number of Sequences: 164201
Number of extensions: 1375210
Number of successful extensions: 4345
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 4315
Number of HSP's gapped (non-prelim): 40
length of query: 348
length of database: 59,974,054
effective HSP length: 111
effective length of query: 237
effective length of database: 41,747,743
effective search space: 9894215091
effective search space used: 9894215091
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146757.11


