

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.3 - phase: 0
(138 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
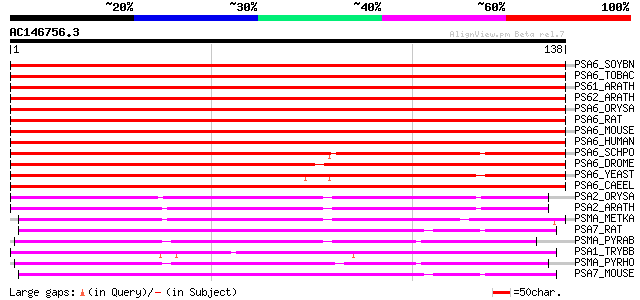
Score E
Sequences producing significant alignments: (bits) Value
PSA6_SOYBN (O48551) Proteasome subunit alpha type 6 (EC 3.4.25.1... 265 2e-71
PSA6_TOBAC (Q9XG77) Proteasome subunit alpha type 6 (EC 3.4.25.1... 252 1e-67
PS61_ARATH (O81146) Proteasome subunit alpha type 6-1 (EC 3.4.25... 248 3e-66
PS62_ARATH (O81147) Proteasome subunit alpha type 6-2 (EC 3.4.25... 244 5e-65
PSA6_ORYSA (Q9LSU3) Proteasome subunit alpha type 6 (EC 3.4.25.1... 236 1e-62
PSA6_RAT (P60901) Proteasome subunit alpha type 6 (EC 3.4.25.1) ... 167 6e-42
PSA6_MOUSE (Q9QUM9) Proteasome subunit alpha type 6 (EC 3.4.25.1... 167 6e-42
PSA6_HUMAN (P60900) Proteasome subunit alpha type 6 (EC 3.4.25.1... 167 6e-42
PSA6_SCHPO (O94517) Probable proteasome subunit alpha type 6 (EC... 150 8e-37
PSA6_DROME (Q9XZJ4) Proteasome subunit alpha type 6 (EC 3.4.25.1... 142 3e-34
PSA6_YEAST (P21243) Proteasome component C7-alpha (EC 3.4.25.1) ... 123 1e-28
PSA6_CAEEL (O17586) Proteasome subunit alpha type 6 (EC 3.4.25.1... 123 1e-28
PSA2_ORYSA (Q9LSU2) Proteasome subunit alpha type 2 (EC 3.4.25.1... 92 3e-19
PSA2_ARATH (O23708) Proteasome subunit alpha type 2 (EC 3.4.25.1... 90 2e-18
PSMA_METKA (Q8TYB7) Proteasome alpha subunit (EC 3.4.25.1) (Mult... 89 2e-18
PSA7_RAT (P48004) Proteasome subunit alpha type 7 (EC 3.4.25.1) ... 86 2e-17
PSMA_PYRAB (Q9V122) Proteasome alpha subunit (EC 3.4.25.1) (Mult... 86 3e-17
PSA1_TRYBB (Q9GU37) Proteasome subunit alpha type 1 (EC 3.4.25.1... 86 3e-17
PSMA_PYRHO (O59219) Proteasome alpha subunit (EC 3.4.25.1) (Mult... 85 4e-17
PSA7_MOUSE (Q9Z2U0) Proteasome subunit alpha type 7 (EC 3.4.25.1... 85 5e-17
>PSA6_SOYBN (O48551) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(20S proteasome alpha subunit A) (20S proteasome subunit
alpha-1) (Proteasome iota subunit)
Length = 246
Score = 265 bits (677), Expect = 2e-71
Identities = 130/138 (94%), Positives = 135/138 (97%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDE GPQLYKCDPAGHYFGHKATSA
Sbjct: 109 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDEYGPQLYKCDPAGHYFGHKATSA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G KDQEAINFLEKKMKNDPSFTY+ETVQTAI+ALQSVLQEDFKATEIEVGVV+KD PEFR
Sbjct: 169 GLKDQEAINFLEKKMKNDPSFTYEETVQTAISALQSVLQEDFKATEIEVGVVRKDNPEFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
VL+T+EIDEHLTAISERD
Sbjct: 229 VLTTDEIDEHLTAISERD 246
>PSA6_TOBAC (Q9XG77) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(20S proteasome alpha subunit A) (20S proteasome subunit
alpha-1)
Length = 246
Score = 252 bits (644), Expect = 1e-67
Identities = 121/138 (87%), Positives = 135/138 (97%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVL+KWIADKSQVYTQHAYMRPLGVVAM+LGID+E GPQL+KCDPAGH+FGHKATSA
Sbjct: 109 MPVDVLSKWIADKSQVYTQHAYMRPLGVVAMILGIDEEKGPQLFKCDPAGHFFGHKATSA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
GSK+QEAINFLEKKMKNDP+F+Y+ETVQTAI+ALQSVLQEDFKA+EIEVGVV+K+ P FR
Sbjct: 169 GSKEQEAINFLEKKMKNDPAFSYEETVQTAISALQSVLQEDFKASEIEVGVVKKEDPIFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
VL+TEEIDEHLTAISERD
Sbjct: 229 VLTTEEIDEHLTAISERD 246
>PS61_ARATH (O81146) Proteasome subunit alpha type 6-1 (EC 3.4.25.1)
(20S proteasome alpha subunit A1)
Length = 246
Score = 248 bits (633), Expect = 3e-66
Identities = 117/138 (84%), Positives = 132/138 (94%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVD+LAKWIADKSQVYTQHAYMRPLGVVAMV+G+D+ENGP LYKCDPAGH++GHKATSA
Sbjct: 109 MPVDILAKWIADKSQVYTQHAYMRPLGVVAMVMGVDEENGPLLYKCDPAGHFYGHKATSA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K+QEA+NFLEKKMK +PSFT+DETVQTAI+ALQSVLQEDFKATEIEVGVV+ + PEFR
Sbjct: 169 GMKEQEAVNFLEKKMKENPSFTFDETVQTAISALQSVLQEDFKATEIEVGVVRAENPEFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
L+TEEI+EHLTAISERD
Sbjct: 229 ALTTEEIEEHLTAISERD 246
>PS62_ARATH (O81147) Proteasome subunit alpha type 6-2 (EC 3.4.25.1)
(20S proteasome alpha subunit A2)
Length = 246
Score = 244 bits (622), Expect = 5e-65
Identities = 118/138 (85%), Positives = 128/138 (92%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MP D+LAKWIADKSQVYTQHAYMRPLGVVAMVLGID+E GP LYKCDPAGH++GHKATSA
Sbjct: 109 MPADILAKWIADKSQVYTQHAYMRPLGVVAMVLGIDEERGPLLYKCDPAGHFYGHKATSA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K+QEA+NFLEKKMK +P+FTYDETVQTAI+ALQSVLQEDFKATEIEVGVV+ D P FR
Sbjct: 169 GMKEQEAVNFLEKKMKENPAFTYDETVQTAISALQSVLQEDFKATEIEVGVVRADDPLFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
L TEEIDEHLTAISERD
Sbjct: 229 SLRTEEIDEHLTAISERD 246
>PSA6_ORYSA (Q9LSU3) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(20S proteasome alpha subunit A) (20S proteasome subunit
alpha-1)
Length = 246
Score = 236 bits (601), Expect = 1e-62
Identities = 115/138 (83%), Positives = 127/138 (91%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVLAKWIADK+QVYTQHAYMRPLGVVAMVLG D+E QL+KCDPAGH+FGHKATSA
Sbjct: 109 MPVDVLAKWIADKAQVYTQHAYMRPLGVVAMVLGYDEEKNAQLFKCDPAGHFFGHKATSA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K+QEAINFLEKKMK+DP F+Y+ETVQ AI+ALQSVLQEDFKATEIEVGVV+KD FR
Sbjct: 169 GLKEQEAINFLEKKMKDDPQFSYEETVQIAISALQSVLQEDFKATEIEVGVVRKDDRVFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
L+TEEID+HLTAISERD
Sbjct: 229 ALTTEEIDQHLTAISERD 246
>PSA6_RAT (P60901) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(Proteasome iota chain) (Macropain iota chain)
(Multicatalytic endopeptidase complex iota chain)
Length = 246
Score = 167 bits (423), Expect = 6e-42
Identities = 81/138 (58%), Positives = 107/138 (76%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PVD+L K IAD SQVYTQ+A MRPLG +++GID+E GPQ+YKCDPAG+Y G KAT+A
Sbjct: 109 IPVDMLCKRIADISQVYTQNAEMRPLGCCMILIGIDEEQGPQVYKCDPAGYYCGFKATAA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K E+ +FLEKK+K +T+++TV+TAI L +VL DFK +EIEVGVV + P+FR
Sbjct: 169 GVKQTESTSFLEKKVKKKFDWTFEQTVETAITCLSTVLSIDFKPSEIEVGVVTVENPKFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
+L+ EID HL A++ERD
Sbjct: 229 ILTEAEIDAHLVALAERD 246
>PSA6_MOUSE (Q9QUM9) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(Proteasome iota chain) (Macropain iota chain)
(Multicatalytic endopeptidase complex iota chain)
Length = 246
Score = 167 bits (423), Expect = 6e-42
Identities = 81/138 (58%), Positives = 107/138 (76%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PVD+L K IAD SQVYTQ+A MRPLG +++GID+E GPQ+YKCDPAG+Y G KAT+A
Sbjct: 109 IPVDMLCKRIADISQVYTQNAEMRPLGCCMILIGIDEEQGPQVYKCDPAGYYCGFKATAA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K E+ +FLEKK+K +T+++TV+TAI L +VL DFK +EIEVGVV + P+FR
Sbjct: 169 GVKQTESTSFLEKKVKKKFDWTFEQTVETAITCLSTVLSIDFKPSEIEVGVVTVENPKFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
+L+ EID HL A++ERD
Sbjct: 229 ILTEAEIDAHLVALAERD 246
>PSA6_HUMAN (P60900) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(Proteasome iota chain) (Macropain iota chain)
(Multicatalytic endopeptidase complex iota chain) (27
kDa prosomal protein) (PROS-27) (p27K)
Length = 246
Score = 167 bits (423), Expect = 6e-42
Identities = 81/138 (58%), Positives = 107/138 (76%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PVD+L K IAD SQVYTQ+A MRPLG +++GID+E GPQ+YKCDPAG+Y G KAT+A
Sbjct: 109 IPVDMLCKRIADISQVYTQNAEMRPLGCCMILIGIDEEQGPQVYKCDPAGYYCGFKATAA 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K E+ +FLEKK+K +T+++TV+TAI L +VL DFK +EIEVGVV + P+FR
Sbjct: 169 GVKQTESTSFLEKKVKKKFDWTFEQTVETAITCLSTVLSIDFKPSEIEVGVVTVENPKFR 228
Query: 121 VLSTEEIDEHLTAISERD 138
+L+ EID HL A++ERD
Sbjct: 229 ILTEAEIDAHLVALAERD 246
>PSA6_SCHPO (O94517) Probable proteasome subunit alpha type 6 (EC
3.4.25.1)
Length = 244
Score = 150 bits (379), Expect = 8e-37
Identities = 81/140 (57%), Positives = 102/140 (72%), Gaps = 4/140 (2%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MP DVLAK +A+ +QV TQ A MRPLGV ++ +DDE GP L+K DPAG Y G+KATSA
Sbjct: 107 MPCDVLAKRMANIAQVSTQRAAMRPLGVAMTLVAVDDEIGPSLFKLDPAGFYIGYKATSA 166
Query: 61 GSKDQEAINFLEKKMKND--PSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPE 118
G K E IN+LEK+ K D PS +TV+T I AL S L DFK+TE+++GVV+ DKP
Sbjct: 167 GPKQTETINWLEKRFKKDGIPS-NLTDTVETGIQALMSSLSTDFKSTELQIGVVEDDKP- 224
Query: 119 FRVLSTEEIDEHLTAISERD 138
FRVLS EEI+ HL +I+E+D
Sbjct: 225 FRVLSVEEIEAHLQSIAEKD 244
>PSA6_DROME (Q9XZJ4) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(20S proteasome subunit alpha-1)
Length = 244
Score = 142 bits (357), Expect = 3e-34
Identities = 73/138 (52%), Positives = 97/138 (69%), Gaps = 2/138 (1%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MPVDVL + IAD +QVYTQ+A MRPLG +++ D+E GP +YK DPAG++ G KA S
Sbjct: 109 MPVDVLCRRIADINQVYTQNAEMRPLGCSMVLIAYDNEIGPSVYKTDPAGYFSGFKACSV 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G+K EA ++LEKK K P+ + ++ +Q AI+ L SVL DFK IE+GVV K P FR
Sbjct: 169 GAKTLEANSYLEKKYK--PNLSEEKAIQLAISCLSSVLAIDFKPNGIEIGVVSKSDPTFR 226
Query: 121 VLSTEEIDEHLTAISERD 138
+L EI+EHLT I+E+D
Sbjct: 227 ILDEREIEEHLTKIAEKD 244
>PSA6_YEAST (P21243) Proteasome component C7-alpha (EC 3.4.25.1)
(Macropain subunit C7-alpha) (Proteinase YSCE subunit 7)
(Multicatalytic endopeptidase complex C7) (Component Y8)
(SCL1 suppressor protein)
Length = 252
Score = 123 bits (309), Expect = 1e-28
Identities = 69/143 (48%), Positives = 92/143 (64%), Gaps = 7/143 (4%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MP DVLAK +A+ SQ+YTQ AYMRPLGV+ + +D+E GP +YK DPAG+Y G+KAT+
Sbjct: 112 MPCDVLAKRMANLSQIYTQRAYMRPLGVILTFVSVDEELGPSIYKTDPAGYYVGYKATAT 171
Query: 61 GSKDQEAINFLE---KKMKND--PSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKD 115
G K QE LE KK K D ++++ V+ AI + L +F ++EVGV KD
Sbjct: 172 GPKQQEITTNLENHFKKSKIDHINEESWEKVVEFAITHMIDALGTEFSKNDLEVGVATKD 231
Query: 116 KPEFRVLSTEEIDEHLTAISERD 138
K F LS E I+E L AI+E+D
Sbjct: 232 K--FFTLSAENIEERLVAIAEQD 252
>PSA6_CAEEL (O17586) Proteasome subunit alpha type 6 (EC 3.4.25.1)
(Proteasome subunit alpha 1)
Length = 246
Score = 123 bits (308), Expect = 1e-28
Identities = 61/138 (44%), Positives = 89/138 (64%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
MP ++LAK +AD +Q YTQ+A MR LG + + DDE GP++Y+ DPAG+Y G K S
Sbjct: 109 MPCELLAKKMADLNQYYTQNAEMRSLGCALLFISYDDEKGPEVYRVDPAGYYRGMKGVSV 168
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G K A +FLEKK+K T E ++ AI ALQ+ L D ++ ++EV VV KD +F
Sbjct: 169 GVKQLPATSFLEKKIKKKSELTSTEAIELAIEALQTSLGIDVRSKDLEVVVVTKDNSKFT 228
Query: 121 VLSTEEIDEHLTAISERD 138
L++++++ HL I+ RD
Sbjct: 229 KLTSDQVEHHLNQIANRD 246
>PSA2_ORYSA (Q9LSU2) Proteasome subunit alpha type 2 (EC 3.4.25.1)
(20S proteasome alpha subunit B) (20S proteasome subunit
alpha-2)
Length = 235
Score = 92.0 bits (227), Expect = 3e-19
Identities = 52/134 (38%), Positives = 75/134 (55%), Gaps = 4/134 (2%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV L + A Q +TQ +RP GV ++ G DD NGPQLY+ DP+G YF KA++
Sbjct: 105 IPVTQLVRETAAVMQEFTQSGGVRPFGVSLLIAGYDD-NGPQLYQVDPSGSYFSWKASAM 163
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G A FLEK+ D D+ + TAI L+ + A IE+G+++ D+ EF+
Sbjct: 164 GKNVSNAKTFLEKRYTED--MELDDAIHTAILTLKEGYEGQISANNIEIGIIRSDR-EFK 220
Query: 121 VLSTEEIDEHLTAI 134
VLS EI + L +
Sbjct: 221 VLSPAEIKDFLEEV 234
>PSA2_ARATH (O23708) Proteasome subunit alpha type 2 (EC 3.4.25.1)
(20S proteasome alpha subunit B)
Length = 235
Score = 89.7 bits (221), Expect = 2e-18
Identities = 52/134 (38%), Positives = 75/134 (55%), Gaps = 4/134 (2%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSA 60
+PV L + A Q +TQ +RP GV +V G DD+ GPQLY+ DP+G YF KA++
Sbjct: 105 IPVTQLVRETATVMQEFTQSGGVRPFGVSLLVAGYDDK-GPQLYQVDPSGSYFSWKASAM 163
Query: 61 GSKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFR 120
G A FLEK+ D D+ + TAI L+ + + + IE+G + DK FR
Sbjct: 164 GKNVSNAKTFLEKRYTED--MELDDAIHTAILTLKEGFEGEISSKNIEIGKIGADK-VFR 220
Query: 121 VLSTEEIDEHLTAI 134
VL+ EID++L +
Sbjct: 221 VLTPAEIDDYLAEV 234
>PSMA_METKA (Q8TYB7) Proteasome alpha subunit (EC 3.4.25.1)
(Multicatalytic endopeptidase complex alpha subunit)
Length = 246
Score = 89.4 bits (220), Expect = 2e-18
Identities = 53/138 (38%), Positives = 79/138 (56%), Gaps = 7/138 (5%)
Query: 3 VDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGS 62
VDVL K I D QVYTQH +RP G ++ G+D + G +L++ DP+G HKAT+ G
Sbjct: 111 VDVLVKAICDLKQVYTQHGGVRPFGTALLIAGVDTK-GCRLFETDPSGALTEHKATAIGE 169
Query: 63 KDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRVL 122
QEA++ E++ + D T E ++ A+ AL +E+ A +E+ VV DK FR L
Sbjct: 170 GRQEALDVFEEEYRED--MTLQEAIELAVRALYEASREETTADNLEIAVV--DKQGFRKL 225
Query: 123 STEEIDEHLTAI--SERD 138
++I+E + SE D
Sbjct: 226 ERKKIEEMFERVVGSEED 243
>PSA7_RAT (P48004) Proteasome subunit alpha type 7 (EC 3.4.25.1)
(Proteasome subunit RC6-1)
Length = 254
Score = 85.9 bits (211), Expect = 2e-17
Identities = 48/134 (35%), Positives = 78/134 (57%), Gaps = 3/134 (2%)
Query: 3 VDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGS 62
V+ + ++IA Q YTQ RP G+ A+++G D + P+LY+ DP+G Y KA + G
Sbjct: 110 VEYITRYIASLKQRYTQSNGRRPFGISALIVGFDFDGTPRLYQTDPSGTYHAWKANAIGR 169
Query: 63 KDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRVL 122
+ FLEK +D T D T++ I AL V+Q K IE+ V+++D+P ++L
Sbjct: 170 GAKSVREFLEKNYTDDAIETDDLTIKLVIKALLEVVQSGGK--NIELAVMRRDQP-LKIL 226
Query: 123 STEEIDEHLTAISE 136
S EEI++++ I +
Sbjct: 227 SPEEIEKYVAEIEK 240
>PSMA_PYRAB (Q9V122) Proteasome alpha subunit (EC 3.4.25.1)
(Multicatalytic endopeptidase complex alpha subunit)
Length = 260
Score = 85.5 bits (210), Expect = 3e-17
Identities = 49/130 (37%), Positives = 76/130 (57%), Gaps = 5/130 (3%)
Query: 2 PVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAG 61
P+ V+ K I D Q++TQ+ +RP G ++ G++D+ P+LY+ DP+G YF KA + G
Sbjct: 110 PLAVIVKKICDLKQMHTQYGGVRPFGAALLMAGVNDK--PELYETDPSGAYFAWKAVAIG 167
Query: 62 SKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRV 121
S A+ E K ++D T DE ++ AI AL +++ A IEV V+ +FR
Sbjct: 168 SGRNTAMAIFEDKYRDD--MTLDEAIKLAIFALAKTMEKP-SAENIEVAVITVKDKKFRK 224
Query: 122 LSTEEIDEHL 131
LS EEI++ L
Sbjct: 225 LSKEEIEKFL 234
>PSA1_TRYBB (Q9GU37) Proteasome subunit alpha type 1 (EC 3.4.25.1)
(20SPA1)
Length = 266
Score = 85.5 bits (210), Expect = 3e-17
Identities = 60/143 (41%), Positives = 79/143 (54%), Gaps = 8/143 (5%)
Query: 1 MPVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGID--DENG---PQLYKCDPAGHYFGH 55
M V VLAK D +QV TQ A MR +G + +G++ DE+G PQ+Y DPAG H
Sbjct: 106 MLVSVLAKPWQDMAQVRTQQAGMRLMGTIMTFVGMEQNDEDGAWIPQIYCVDPAGGAAVH 165
Query: 56 KATSAGSKDQEAINFLEKKMKNDPSFTYD--ETVQTAIAALQSVLQEDFKATEIEVGVVQ 113
A + G K EA FLEKK KN P T E A+AALQS L E +A+ +EVG
Sbjct: 166 -ACAVGKKQIEACAFLEKKQKNAPFHTLSQKEAAMIALAALQSALGESLRASGVEVGRCT 224
Query: 114 KDKPEFRVLSTEEIDEHLTAISE 136
D F +S E+ + LT +++
Sbjct: 225 ADDHHFLRVSDREVKKWLTPLAK 247
>PSMA_PYRHO (O59219) Proteasome alpha subunit (EC 3.4.25.1)
(Multicatalytic endopeptidase complex alpha subunit)
Length = 260
Score = 85.1 bits (209), Expect = 4e-17
Identities = 47/133 (35%), Positives = 79/133 (59%), Gaps = 5/133 (3%)
Query: 2 PVDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAG 61
P+ V+ K I D Q++TQ+ +RP G ++ G++++ P+LY+ DP+G YF KA + G
Sbjct: 110 PLAVIVKKICDLKQMHTQYGGVRPFGAALLMAGVNEK--PELYETDPSGAYFAWKAVAIG 167
Query: 62 SKDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRV 121
S A+ E+K K+D S +E ++ AI AL +++ A IEV ++ +FR
Sbjct: 168 SGRNTAMAIFEEKYKDDMSL--EEAIKLAIFALAKTMEKP-SAENIEVAIITVKDKKFRK 224
Query: 122 LSTEEIDEHLTAI 134
LS EEI+++L +
Sbjct: 225 LSREEIEKYLNEV 237
>PSA7_MOUSE (Q9Z2U0) Proteasome subunit alpha type 7 (EC 3.4.25.1)
(Proteasome subunit RC6-1)
Length = 248
Score = 84.7 bits (208), Expect = 5e-17
Identities = 47/134 (35%), Positives = 78/134 (58%), Gaps = 3/134 (2%)
Query: 3 VDVLAKWIADKSQVYTQHAYMRPLGVVAMVLGIDDENGPQLYKCDPAGHYFGHKATSAGS 62
V+ + ++IA Q YTQ RP G+ A+++G D + P+LY+ DP+G Y KA + G
Sbjct: 104 VEYITRYIASLKQRYTQSNGRRPFGISALIVGFDFDGTPRLYQTDPSGTYHAWKANAIGR 163
Query: 63 KDQEAINFLEKKMKNDPSFTYDETVQTAIAALQSVLQEDFKATEIEVGVVQKDKPEFRVL 122
+ FLEK +D T D T++ I AL V+Q K IE+ V+++D+P ++L
Sbjct: 164 GAKSVREFLEKNYTDDAIETDDLTIKLVIKALLEVVQSGGK--NIELAVMRRDQP-LKIL 220
Query: 123 STEEIDEHLTAISE 136
+ EEI++++ I +
Sbjct: 221 NPEEIEKYVAEIEK 234
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,277,591
Number of Sequences: 164201
Number of extensions: 635269
Number of successful extensions: 1650
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 131
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1432
Number of HSP's gapped (non-prelim): 155
length of query: 138
length of database: 59,974,054
effective HSP length: 99
effective length of query: 39
effective length of database: 43,718,155
effective search space: 1705008045
effective search space used: 1705008045
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146756.3


