

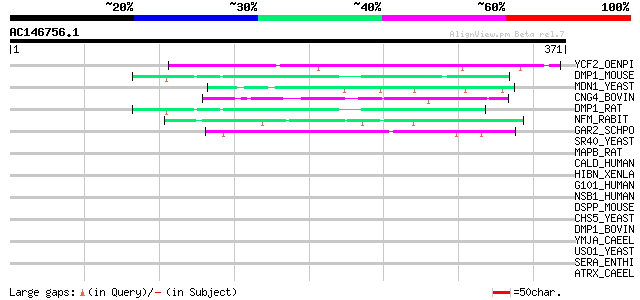
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146756.1 - phase: 0
(371 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YCF2_OENPI (P31568) Protein ycf2 (Fragment) 49 2e-05
DMP1_MOUSE (O55188) Dentin matrix acidic phosphoprotein 1 precur... 48 5e-05
MDN1_YEAST (Q12019) Midasin (MIDAS-containing protein) 47 1e-04
CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor CNG-cha... 46 1e-04
DMP1_RAT (P98193) Dentin matrix acidic phosphoprotein 1 precurso... 46 2e-04
NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa neur... 44 5e-04
GAR2_SCHPO (P41891) Protein gar2 44 5e-04
SR40_YEAST (P32583) Suppressor protein SRP40 43 0.001
MAPB_RAT (P15205) Microtubule-associated protein 1B (MAP 1B) (Ne... 43 0.001
CALD_HUMAN (Q05682) Caldesmon (CDM) 42 0.002
HIBN_XENLA (P06180) Histone-binding protein N1/N2 42 0.002
G101_HUMAN (Q96P66) Probable G protein-coupled receptor GPR101 42 0.002
NSB1_HUMAN (P82970) Nucleosomal binding protein 1 42 0.003
DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin... 41 0.006
CHS5_YEAST (Q12114) Chitin biosynthesis protein CHS5 (CAL3 protein) 40 0.009
DMP1_BOVIN (Q95120) Dentin matrix acidic phosphoprotein 1 precur... 40 0.012
YMJA_CAEEL (P34486) Hypothetical protein F59B2.11 in chromosome III 39 0.016
USO1_YEAST (P25386) Intracellular protein transport protein USO1 39 0.016
SERA_ENTHI (P21138) Serine-rich 25 kDa antigen protein (SHEHP) (... 39 0.016
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 39 0.016
>YCF2_OENPI (P31568) Protein ycf2 (Fragment)
Length = 721
Score = 48.9 bits (115), Expect = 2e-05
Identities = 58/272 (21%), Positives = 115/272 (41%), Gaps = 15/272 (5%)
Query: 107 KQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVE-TEPDLAVSGEGRLPENEK 165
++ V +V+ ++ + + +V+ EDE+ + EE+VE TE ++ + E E+
Sbjct: 183 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 242
Query: 166 TGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSV--NESNSTGSRFDALKTGEV 223
G DE V DE T++ + EE + + V E G+ + ++ E
Sbjct: 243 VEGTEDEEVEGTE--DEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEE 300
Query: 224 DVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSE 283
+V+ G+ +V+ E + T + + E+E G+ + E E
Sbjct: 301 EVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEV 360
Query: 284 ERKVEDDSVTRSEGGGTR---ESSEVQSSSSLSKTRRTQRRRKEVSGED-AKMENEDVAV 339
E E+ T E GT E +E + + + T+ E + +D ++ +N+ V +
Sbjct: 361 EGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEKDSSQFDNDRVTL 420
Query: 340 V---KSEPLFGVLEMIKRHQKFSLFERRLEKN 368
+ K + +I +HQK +E LE++
Sbjct: 421 LLRPKPRNPLDIQRLIYQHQK---YESELEED 449
>DMP1_MOUSE (O55188) Dentin matrix acidic phosphoprotein 1 precursor
(Dentin matrix protein-1) (DMP-1) (AG1)
Length = 503
Score = 47.8 bits (112), Expect = 5e-05
Identities = 56/258 (21%), Positives = 100/258 (38%), Gaps = 26/258 (10%)
Query: 83 QQNGDVSAVTAEDSDHVPWLD-----KLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDE 137
+ GD + +DSD ++ R+ RV+E +L+D + S + Q ED
Sbjct: 236 ESKGDREPTSTQDSDDSQSVEFSSRKSFRRSRVSEEDYRGELTDSN--SRETQSDSTEDT 293
Query: 138 KAKENEEKVETEPDLAVS-GEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGE 196
+KE E + E++ D A S + PE + + E + S + + G+
Sbjct: 294 ASKE-ESRSESQEDTAESQSQEDSPEGQDPSSESSEEAGEPSQESSSESQEGVTSESRGD 352
Query: 197 ESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKS 256
D +Q+ ++ +S S D+L T S + +S + D + S
Sbjct: 353 NPDNTSQTGDQEDSESSEEDSLNT--------------FSSSESQSTEEQADSESNESLS 398
Query: 257 VEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTR 316
+ EES + D + + +S S E + ++ ++SE E SS SK
Sbjct: 399 LSEESQESAQDGDSSSQEGLQSQSASTESRSQE---SQSEQDSRSEEDSDSQDSSRSKEE 455
Query: 317 RTQRRRKEVSGEDAKMEN 334
S ED + +N
Sbjct: 456 SNSTGSASSSEEDIRPKN 473
Score = 34.7 bits (78), Expect = 0.40
Identities = 46/218 (21%), Positives = 84/218 (38%), Gaps = 21/218 (9%)
Query: 136 DEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAG 195
D K +++E+ + PD S + E ++ GG + + + D+ + +
Sbjct: 159 DSKDQDSEDDAHSRPDAGDSAQHSESEEQRVGGGSE----GQSSHGDGSEFDDEGMQSDD 214
Query: 196 EESDRDNQSVNESNSTGSRFDALK-----TGEVDVKLGSGPGHGSGSVQVKSGLNEPDQT 250
ES R ++ +S G R + K T D S +S ++E D
Sbjct: 215 PESTRSDRGHARMSSAGIRSEESKGDREPTSTQDSDDSQSVEFSSRKSFRRSRVSEEDYR 274
Query: 251 GRKRKSVEEESNNGSYDN--------NEAKAVTCESVPPSEERKVEDDSVTRSEGGG--T 300
G S E+ + S ++ +E++ T ES + + +D S SE G +
Sbjct: 275 GELTDSNSRETQSDSTEDTASKEESRSESQEDTAESQSQEDSPEGQDPSSESSEEAGEPS 334
Query: 301 RESSEVQSSSSLSKTRRTQRRRKEVSG--EDAKMENED 336
+ESS S++R +G ED++ ED
Sbjct: 335 QESSSESQEGVTSESRGDNPDNTSQTGDQEDSESSEED 372
>MDN1_YEAST (Q12019) Midasin (MIDAS-containing protein)
Length = 4910
Score = 46.6 bits (109), Expect = 1e-04
Identities = 53/242 (21%), Positives = 94/242 (37%), Gaps = 45/242 (18%)
Query: 133 KLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLP 192
K+ D+KA +N ++ +T+ +L +G+ E + + DE +R ++ + D P
Sbjct: 4158 KMWDDKASDNSKEKDTDQNL----DGKNQEEDVQAAENDEQ----QRDNKEGGDEDPNAP 4209
Query: 193 AAGEESDRDNQSVNESNSTGSRFDALKTGE-----------------VDVKLGSGPGHGS 235
G+E ++++ E N G + D +K E D+ L S
Sbjct: 4210 EDGDEEIENDENAEEENDVGEQEDEVKDEEGEDLEANVPEIETLDLPEDMNLDSEHEESD 4269
Query: 236 GSVQVKSGLNE---PDQTGRKRKSVEEESNNGSYDNN-----EAKAVTCESVPPSEERKV 287
V + G+ + ++ G + + V++ES S + N E A E+ EE
Sbjct: 4270 EDVDMSDGMPDDLNKEEVGNEDEEVKQESGIESDNENDEPGPEEDAGETETALDEEEGAE 4329
Query: 288 EDDSVTRSEGGGTRES---SEVQSSSSLSKTRRTQRRRKEVSGE---------DAKMENE 335
ED +T EG E+ + S K KE GE + K + E
Sbjct: 4330 EDVDMTNDEGKEDEENGPEEQAMSDEEELKQDAAMEENKEKGGEQNTEGLDGVEEKADTE 4389
Query: 336 DV 337
D+
Sbjct: 4390 DI 4391
Score = 39.3 bits (90), Expect = 0.016
Identities = 43/215 (20%), Positives = 87/215 (40%), Gaps = 19/215 (8%)
Query: 135 EDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGD--------IDEPVPAIRRLDESTTN 186
EDE+ E+ + E +L + + EN++ GG+ ++E E+
Sbjct: 4341 EDEENGPEEQAMSDEEEL--KQDAAMEENKEKGGEQNTEGLDGVEEKADTEDIDQEAAVQ 4398
Query: 187 TDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNE 246
D AG ++ D Q ++ +G+ + + + DV + + +K +
Sbjct: 4399 QDSGSKGAGADAT-DTQEQDDVGGSGTTQNTYEEDQEDVTKNNEESREEATAALKQLGDS 4457
Query: 247 PDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVE---DDSVTRSEGGGTRES 303
+ R+R+ ++E NG D N K + P E VE ++ T++ G T++
Sbjct: 4458 MKEYHRRRQDIKEAQTNGEEDENLEK----NNERPDEFEHVEGANTETDTQALGSATQDQ 4513
Query: 304 SE-VQSSSSLSKTRRTQRRRKEVSGEDAKMENEDV 337
+ + ++ R Q ++ EDA E D+
Sbjct: 4514 LQTIDEDMAIDDDREEQEVDQKELVEDADDEKMDI 4548
>CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor
CNG-channel [Contains: Glutamic acid-rich protein
(GARP); Cyclic-nucleotide-gated cation channel 4 (CNG
channel 4) (CNG-4) (Cyclic nucleotide-gated cation
channel modulatory subunit)]
Length = 1394
Score = 46.2 bits (108), Expect = 1e-04
Identities = 50/207 (24%), Positives = 88/207 (42%), Gaps = 29/207 (14%)
Query: 130 QVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDK 189
++ ++++EK E EEK + E + EGR E E+ G+ E + +E
Sbjct: 349 ELPRIQEEKEDEEEEKEDGEEE---EEEGR--EKEEEEGEEKEEEEGREKEEEE------ 397
Query: 190 LLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQ 249
GE+ + + + E + + E + G G + + G E ++
Sbjct: 398 -----GEKKEEEGREKEEEEGGEKEDEEGREKEEE--------EGRGKEEEEGGEKEEEE 444
Query: 250 TGRKRKSVEEESNNGSYDNNEAKAVTCES--VPPSEE-RKVEDDSVTRSEGGGTRESSEV 306
GR ++ VE + + +V +S VP SEE R E ++ +SE GG + EV
Sbjct: 445 -GRGKEEVEGREEEEDEEEEQDHSVLLDSYLVPQSEEDRSEESETQDQSEVGGAQAQGEV 503
Query: 307 QSSSSLSKTRRTQRRRKEVSGEDAKME 333
+ +LS+ TQ + EV G + E
Sbjct: 504 GGAQALSEESETQ-DQSEVGGAQDQSE 529
>DMP1_RAT (P98193) Dentin matrix acidic phosphoprotein 1 precursor
(Dentin matrix protein-1) (DMP-1) (AG1)
Length = 489
Score = 45.8 bits (107), Expect = 2e-04
Identities = 55/243 (22%), Positives = 94/243 (38%), Gaps = 24/243 (9%)
Query: 83 QQNGDVSAVTAEDSDHVPWLD-----KLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDE 137
+ GD + +DSD ++ R+ RV+E +L+D + S + Q ED
Sbjct: 220 ESKGDHEPTSTQDSDDSQDVEFSSRKSFRRSRVSEEDDRGELADSN--SRETQSVSTEDF 277
Query: 138 KAKENEEKVETEPDLA-VSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGE 196
++KE E + ET+ D A + PE + + E + S + + G+
Sbjct: 278 RSKE-ESRSETQEDTAETQSQEDSPEGQDPSSESSEEAGEPSQESSSESQEGVASESRGD 336
Query: 197 ESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKS 256
D +Q+ ++ +S S D L T S + +S + D + S
Sbjct: 337 NPDNTSQTGDQRDSESSEEDRLNT--------------FSSSESQSTEEQGDSESNESLS 382
Query: 257 VEEESNNGSYDNNEAKAVTCESVPPS-EERKVEDDSVTRSEGGGTRESSEVQSSSSLSKT 315
+ EES + D + + +S S E R E S S R+S SS S ++
Sbjct: 383 LSEESQESAQDEDSSSQEGLQSQSASRESRSQESQSEQDSRSEENRDSDSQDSSRSKEES 442
Query: 316 RRT 318
T
Sbjct: 443 NST 445
>NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Fragment)
Length = 644
Score = 44.3 bits (103), Expect = 5e-04
Identities = 57/251 (22%), Positives = 99/251 (38%), Gaps = 18/251 (7%)
Query: 104 KLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPEN 163
K++K +V + VQ V + ++ K+EDEK++ + +LAVS + E
Sbjct: 221 KIQKTKVEAPKLKVQHKFVEEI---IEETKVEDEKSEMEDALTAIAEELAVSVKEEEKEE 277
Query: 164 EKTG----GDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALK 219
E G + +E V A ++ T T ++ GE+ + + E G + D +
Sbjct: 278 EAEGKEEEQEAEEEVAAAKKSPVKAT-TPEIKEEEGEKEEEGQEEEEEEEDEGVKSDQAE 336
Query: 220 TGEVDVKLGSGPGHG---SGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDN---NEAKA 273
G + K GS G G + + + E + ++ K EE+S + EAK
Sbjct: 337 EGGSE-KEGSSKNEGEQEEGETEAEGEVEEAE--AKEEKKTEEKSEEVAAKEEPVTEAKV 393
Query: 274 VTCESV-PPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKM 332
E P + VE+ G E E + K + + KE E +
Sbjct: 394 GKPEKAKSPVPKSPVEEVKPKAEATAGKGEQKEEEEKVEEEKKKAAKESPKEEKVEKKEE 453
Query: 333 ENEDVAVVKSE 343
+ +DV K+E
Sbjct: 454 KPKDVPKKKAE 464
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 44.3 bits (103), Expect = 5e-04
Identities = 45/217 (20%), Positives = 91/217 (41%), Gaps = 12/217 (5%)
Query: 132 KKLEDEKAKE---NEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTD 188
KK+ E AKE K + P + R E + + + + ++ + S+ +
Sbjct: 30 KKITKEAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQKKSKKKEESSSESES 89
Query: 189 KLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPD 248
+ + E S +++S + + + S + E +V + + S S S +E +
Sbjct: 90 ESSSSESESSSSESESSSSESESSSSESSSSESEEEVIVKTEEKKESSSESSSSSESEEE 149
Query: 249 QTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEG---GGTRESSE 305
+ + +EE+ + S ++E+ + ES S E + E++ V ++E G + SS+
Sbjct: 150 EEAVVK--IEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSESSSD 207
Query: 306 VQSSSSLSK----TRRTQRRRKEVSGEDAKMENEDVA 338
+SSS S + + E S ED K + A
Sbjct: 208 SESSSDSSSESGDSDSSSDSESESSSEDEKKRKAEPA 244
Score = 32.0 bits (71), Expect = 2.6
Identities = 35/175 (20%), Positives = 72/175 (41%), Gaps = 8/175 (4%)
Query: 176 AIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKL---GSGPG 232
AI + +S T + ++S + + S +S R + + + VK
Sbjct: 22 AIEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQKKSKKKE 81
Query: 233 HGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSV 292
S + +S +E + + + +S ES + S +++ +++ V E+++ +S
Sbjct: 82 ESSSESESESSSSESESSSSESESSSSESESSSSESSSSESEEEVIVKTEEKKESSSESS 141
Query: 293 TRSEGGGTRES----SEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVKSE 343
+ SE E+ E + SSS S + + + S E E+V V K+E
Sbjct: 142 SSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEEV-VEKTE 195
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 43.1 bits (100), Expect = 0.001
Identities = 42/197 (21%), Positives = 73/197 (36%), Gaps = 7/197 (3%)
Query: 138 KAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEE 197
K KE EEK + + S + + E + S++++ + +
Sbjct: 17 KEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSS----SSSSSSSSSSDSSDSSD 72
Query: 198 SDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSV 257
S+ + S + S+S+ S D+ + E D S S S +E + +KR
Sbjct: 73 SESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKR--A 130
Query: 258 EEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRR 317
E N + + +AK ES SE S + SE G +S SSSS S +
Sbjct: 131 RESDNEDAKETKKAKTEP-ESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSES 189
Query: 318 TQRRRKEVSGEDAKMEN 334
+ S + ++
Sbjct: 190 DSESDSQSSSSSSSSDS 206
Score = 40.4 bits (93), Expect = 0.007
Identities = 45/209 (21%), Positives = 78/209 (36%), Gaps = 15/209 (7%)
Query: 136 DEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAG 195
DE + E+E + ET+ S E +K EP + S+ ++ +G
Sbjct: 114 DESSSESESEDETKKRARESDNEDAKETKKAK---TEPESSSSSESSSSGSSSSSESESG 170
Query: 196 EESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRK 255
ESD D+ S + S+S D+ E D + S S S + D +
Sbjct: 171 SESDSDSSSSSSSSS-----DSESDSESDSQSSSSSSSSDSSSD--SDSSSSDSSSDSDS 223
Query: 256 SVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKT 315
S S++ D++ + +S S+ D S S + +S S S S
Sbjct: 224 SSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDSDSDSGSSS-- 281
Query: 316 RRTQRRRKEVSGEDAKMENEDVAVVKSEP 344
+ KE + +++K E + +S P
Sbjct: 282 ---ELETKEATADESKAEETPASSNESTP 307
Score = 39.7 bits (91), Expect = 0.012
Identities = 38/232 (16%), Positives = 87/232 (37%), Gaps = 16/232 (6%)
Query: 118 QLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAI 177
++ +S+ +++ K + + + + S G + + +
Sbjct: 10 EVPKLSVKEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSD 69
Query: 178 RRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTG---------EVDVKLG 228
ES++++ ++ SD ++ S ++S+S+GS + + E + K
Sbjct: 70 SSDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKR 129
Query: 229 SGPGHGSGSVQVKSGLNEPDQTGRKRKSVE-----EESNNGSYDNNEAKAVTCESVPPSE 283
+ + + K EP+ + S ES +GS ++++ + + S
Sbjct: 130 ARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSS--SDS 187
Query: 284 ERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENE 335
E E DS + S + SS+ SSSS S + S D+ +++
Sbjct: 188 ESDSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSD 239
Score = 32.7 bits (73), Expect = 1.5
Identities = 28/128 (21%), Positives = 47/128 (35%), Gaps = 3/128 (2%)
Query: 217 ALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQT-GRKRKSVEEESNNGSYDNNEAK--A 273
++K E++ K S S S S + + G S S++ S D++++
Sbjct: 15 SVKEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSE 74
Query: 274 VTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKME 333
+ S S DS + SE + S SSSS ++ E + +
Sbjct: 75 SSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESD 134
Query: 334 NEDVAVVK 341
NED K
Sbjct: 135 NEDAKETK 142
>MAPB_RAT (P15205) Microtubule-associated protein 1B (MAP 1B)
(Neuraxin) [Contains: MAP1 light chain LC1]
Length = 2459
Score = 43.1 bits (100), Expect = 0.001
Identities = 67/298 (22%), Positives = 120/298 (39%), Gaps = 40/298 (13%)
Query: 70 DLNRRFKDDVPPPQQNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQL 129
+L + K + P +V ++ V +K K+ + ++ +D++ S + LS
Sbjct: 692 ELKKEVKKETPLKDAKKEVKK---DEKKEVKKEEKEPKKEIKKISKDIKKS--TPLSDTK 746
Query: 130 QVKKLEDEKAKENEEKVETEPDLA--VSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNT 187
+ L+ + AK+ EE + EP A + +G++ +K G + A+ T
Sbjct: 747 KPAALKPKVAKK-EEPTKKEPIAAGKLKDKGKVKVIKKEGKTTEAAATAV--------GT 797
Query: 188 DKLLPAAG-------EESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQV 240
+ AAG +E + + ++ F+ LK E+DV P +
Sbjct: 798 AAVAAAAGVAASGPAKELEAERSLMSSPEDLTKDFEELKAEEIDVAKDIKPQLELIEDEE 857
Query: 241 KSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVT------CESVP----PSEERKVEDD 290
K EP + +K E E + GS ++ + T CE P P E++ V+D
Sbjct: 858 KLKETEPGEAYVIQK--ETEVSKGSAESPDEGITTTEGEGECEQTPEELEPVEKQGVDDI 915
Query: 291 SVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKE----VSGEDAK-MENEDVAVVKSE 343
EG G ESSE ++T + ++ VSG +K ED + K+E
Sbjct: 916 EKFEDEGAGFEESSEAGDYEEKAETEEAEEPEEDGEDNVSGSASKHSPTEDEEIAKAE 973
>CALD_HUMAN (Q05682) Caldesmon (CDM)
Length = 793
Score = 42.4 bits (98), Expect = 0.002
Identities = 50/260 (19%), Positives = 105/260 (40%), Gaps = 17/260 (6%)
Query: 94 EDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVK---KLEDEKAKENEEKVETEP 150
+D + ++ R+ R LR+ + + ++ Q++V + DE+AK + E
Sbjct: 31 DDDEEEAARERRRRARQERLRQKQEEESLGQVTDQVEVNAQNSVPDEEAKTTTTNTQVEG 90
Query: 151 DLAVSGEGRLPENEKTGGD-IDEPVPAIRRLDESTTNTDKLLPAAGEESDR-DNQSVNES 208
D + RL E+ + E + + D + T+ LP+ ++D +N++ +
Sbjct: 91 DDEAAFLERLARREERRQKRLQEALERQKEFDPTITDASLSLPSRRMQNDTAENETTEKE 150
Query: 209 NSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDN 268
+ SR + + E + S + + + K + EE+ GS
Sbjct: 151 EKSESRQERYEIEETETVTKSYQKNDWRDAE-----ENKKEDKEKEEEEEEKPKRGSIGE 205
Query: 269 NEAKAVTCESVPPSEERKV----EDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKE 324
N+ + + E S+E V ++ ++ E +E Q S +S + + KE
Sbjct: 206 NQVEVMVEEKTTESQEETVVMSLKNGQISSEE--PKQEEEREQGSDEISHHEKMEEEDKE 263
Query: 325 -VSGEDAKMENEDVAVVKSE 343
E A++E E+ +K+E
Sbjct: 264 RAEAERARLEAEERERIKAE 283
Score = 33.9 bits (76), Expect = 0.68
Identities = 58/277 (20%), Positives = 109/277 (38%), Gaps = 34/277 (12%)
Query: 104 KLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPEN 163
K ++R AE R+ ++ + + + + E+EKAK E+K + + + R +
Sbjct: 346 KEEEKRAAEERQRIKEEEKRAAEERQRARAEEEEKAKVEEQKRNKQ----LEEKKRAMQE 401
Query: 164 EKTGGDIDEPVPAIRRLDESTTNTDKL----LPAAGEESDRDNQSVNES-NSTGSRFDAL 218
K G+ E + ++E DKL L GEE Q+ E F
Sbjct: 402 TKIKGEKVEQKIEGKWVNEKKAQEDKLQTAVLKKQGEEKGTKVQAKREKLQEDKPTFKKE 461
Query: 219 KTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCES 278
+ + +K P +VKS ++ RK+ E +S NG + ++ K
Sbjct: 462 EIKDEKIKKDKEP-----KEEVKSFMD------RKKGFTEVKSQNGEFMTHKLKHTENTF 510
Query: 279 VPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVA 338
P V+ +EG + +V++ L + RR + + E K + ++ A
Sbjct: 511 SRPGGRASVD---TKEAEG-----APQVEAGKRLEELRRRRGETESEEFEKLKQKQQEAA 562
Query: 339 V------VKSEPLFGVLEMIKRHQKFSLFERRLEKNQ 369
+ K E VLE ++ +K +R+L + +
Sbjct: 563 LELEELKKKREERRKVLEEEEQRRKQEEADRKLREEE 599
>HIBN_XENLA (P06180) Histone-binding protein N1/N2
Length = 589
Score = 42.0 bits (97), Expect = 0.002
Identities = 48/207 (23%), Positives = 87/207 (41%), Gaps = 15/207 (7%)
Query: 140 KENEEKVETEPDLAVSGEGRLPENEKTG--GDIDEPVPAIRRLDESTTNTDKLLPAAGEE 197
+++EE+ E E D + L E E+ + + + +R + T+ ++ G+
Sbjct: 107 EDDEEEAEKEEDPNIPSADNLDEKEREQLREQVYDAMAEDQRAPDDTSESEAKGKPEGDS 166
Query: 198 SDR--DNQSVNESNSTGSRFDALKTGEV--DV---KLGSGPGHGSGSVQVKSGLNEPDQT 250
D+ D + N T D LK DV K G G S + E
Sbjct: 167 KDKEADEKMKNGQKETEKVTDDLKIDSASRDVPMDKSGKGEPPESKDAETLVEQKESKPE 226
Query: 251 GRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEV-QSS 309
K KS+E + + S + +AK +S +E + + D SE ++ES+ + +
Sbjct: 227 TLKEKSIETKEKDLSKEKTDAKETANQSPDSTEVAEEKMD----SEASESKESTSIPPTE 282
Query: 310 SSLSKTRRTQRRRKEVSGEDAKMENED 336
+ +K ++ +E GED++ ENED
Sbjct: 283 NEANKPDDPEKMEEEEEGEDSE-ENED 308
>G101_HUMAN (Q96P66) Probable G protein-coupled receptor GPR101
Length = 508
Score = 42.0 bits (97), Expect = 0.002
Identities = 46/187 (24%), Positives = 79/187 (41%), Gaps = 16/187 (8%)
Query: 111 AELRRDVQLSDVSILSLQLQVKKL---EDEKAKENEEKVETEPDLAVSGEGRLPENEKTG 167
A R+ L +V SL+++VK EDE+ E +E+ + E + EG + E
Sbjct: 218 AARRQHALLYNVKRHSLEVRVKDCVENEDEEGAEKKEEFQDESEFRRQHEGEVKAKE--- 274
Query: 168 GDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKL 227
G ++ +++ + ST ++ + A G E R++ +V S + + K E +K
Sbjct: 275 GRMEAKDGSLKAKEGSTGTSESSVEARGSEEVRESSTVASDGSMEGKEGSTKVEENSMKA 334
Query: 228 GSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKV 287
G + Q L E D E+ N S D+ EA + ES+PPS
Sbjct: 335 DKGRTEVN---QCSIDLGEDDM------EFGEDDINFSEDDVEAVNIP-ESLPPSRRNSN 384
Query: 288 EDDSVTR 294
+ + R
Sbjct: 385 SNPPLPR 391
>NSB1_HUMAN (P82970) Nucleosomal binding protein 1
Length = 282
Score = 41.6 bits (96), Expect = 0.003
Identities = 53/246 (21%), Positives = 89/246 (35%), Gaps = 36/246 (14%)
Query: 107 KQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEP----DLAVSGEGRLPE 162
K + E D V+ + V++ +E AK E K+ P ++ E + +
Sbjct: 48 KSDMMEENIDTSAQAVAETKQEAVVEEDYNENAKNGEAKITEAPASEKEIVEVKEENIED 107
Query: 163 NEKTGGDIDEPVPAI-------RRLDESTTNTDKLLPAA------GEESDRDNQSVNESN 209
+ GG+ E V A ++ DE N +K GEE +++++ NE
Sbjct: 108 ATEKGGEKKEAVAAEVKNEEEDQKEDEEDQNEEKGEAGKEDKDEKGEEDGKEDKNGNEKG 167
Query: 210 STGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSY--D 267
+ K GE G G+G + G +E ++ RK +E+ +G D
Sbjct: 168 EDAKEKEDGKKGE--------DGKGNGEDGKEKGEDEKEEEDRKETGDGKENEDGKEKGD 219
Query: 268 NNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSG 327
E K V + E ED+ E G +E K + E+
Sbjct: 220 KKEGKDVKVKEDEKEREDGKEDEGGNEEEAGKEKED---------LKEEEEGKEEDEIKE 270
Query: 328 EDAKME 333
+D K E
Sbjct: 271 DDGKKE 276
>DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin
matrix protein-3) (DMP-3) [Contains: Dentin
phosphoprotein (Dentin phosphophoryn) (DPP); Dentin
sialoprotein (DSP)]
Length = 934
Score = 40.8 bits (94), Expect = 0.006
Identities = 36/144 (25%), Positives = 63/144 (43%), Gaps = 5/144 (3%)
Query: 196 EESDRDNQSVNESNSTG-SRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKR 254
+ESD +++S NES S G + + + ++ + D S G S SG + D G
Sbjct: 464 DESDTNSESANESGSRGDASYTSDESSDDDNDSDSHAGEDDSSDD-SSGDGDSDSNGDGD 522
Query: 255 KSVEE--ESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSL 312
E+ ES++ +DN+ +S S++ DS S+ + +SS+ S SS
Sbjct: 523 SESEDKDESDSSDHDNSSDSESKSDSSDSSDDSSDSSDSSDSSDSSDSSDSSD-SSDSSD 581
Query: 313 SKTRRTQRRRKEVSGEDAKMENED 336
S + + SG ++ D
Sbjct: 582 SSDSNSSSDSSDSSGSSDSSDSSD 605
>CHS5_YEAST (Q12114) Chitin biosynthesis protein CHS5 (CAL3 protein)
Length = 671
Score = 40.0 bits (92), Expect = 0.009
Identities = 42/201 (20%), Positives = 86/201 (41%), Gaps = 21/201 (10%)
Query: 136 DEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVP----AIRRLDESTTNTDKLL 191
+E A+++ E VE + ++ K D ++PV + +E +T + +
Sbjct: 475 NEPAEDSNEPVEDSNKPVKDSNKPVEDSNKPVEDSNKPVEDSNKPVEDANEPVEDTSEPV 534
Query: 192 PAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTG 251
AGE V E+N + + + E D++L + P + SV V+ NE +
Sbjct: 535 EDAGEP-------VQETNEFTTDIASPRHQEEDIELEAEPKDATESVAVEPS-NEDVKPE 586
Query: 252 RKRKSVEEESNNGSYDNNEAKAVT---------CESVPPSEERKVEDDSVTRSEGGGTRE 302
K E++ NN S + ++ T ES +EE++ + V + T+E
Sbjct: 587 EKGSEAEDDINNVSKEAASGESTTHQKTEASASLESSAVTEEQETTEAEVNTDDVLSTKE 646
Query: 303 SSEVQSSSSLSKTRRTQRRRK 323
+ + +S+ +K + + ++K
Sbjct: 647 AKKNTGNSNSNKKKNKKNKKK 667
>DMP1_BOVIN (Q95120) Dentin matrix acidic phosphoprotein 1 precursor
(Dentin matrix protein-1) (DMP-1)
Length = 510
Score = 39.7 bits (91), Expect = 0.012
Identities = 56/243 (23%), Positives = 96/243 (39%), Gaps = 20/243 (8%)
Query: 114 RRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETE----PDLAVSGEGRLPENEKTGGD 169
R + ++SD + S Q K +DE+ ++ E+ P + RLPE E G+
Sbjct: 227 RGNSRISDAGLKSTQ---SKGDDEEQASTQDSHESPAAAYPRRKFFRKSRLPE-EDGRGE 282
Query: 170 IDEPVPAIRRLDESTTNTDKLLPAAGEESDRD-NQSVNESNSTGSRFDAL--------KT 220
+D+ I + +ST N D G+ + ++S ES S D+ +
Sbjct: 283 LDDS-RTIEVMSDSTENPDSKEAGLGQSREHSKSESRQESEENRSPEDSQDVQDPSSESS 341
Query: 221 GEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVP 280
EVD+ +S + PD + + +S + D + K ES
Sbjct: 342 QEVDLPSQENSSESQEEALHESRGDNPDNATSHSREHQADSESSEEDVLD-KPSDSESTS 400
Query: 281 PSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVV 340
E+ E RS ES+E Q+SSS + R ++ S ED +++D +
Sbjct: 401 TEEQADSESHESLRSSEESP-ESTEEQNSSSQEGAQTQSRSQESPSEEDDGSDSQDSSRS 459
Query: 341 KSE 343
K +
Sbjct: 460 KED 462
>YMJA_CAEEL (P34486) Hypothetical protein F59B2.11 in chromosome III
Length = 340
Score = 39.3 bits (90), Expect = 0.016
Identities = 45/251 (17%), Positives = 99/251 (38%), Gaps = 21/251 (8%)
Query: 88 VSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSD--VSILSLQLQVKKLEDEKAKENEEK 145
V +D+ W K+ ++ A R++ SD +++ L++ +K +E
Sbjct: 38 VGVKMGQDAVEGNWKRKIIPKKTAI--REILKSDEEITVRHLKIGLKSRRLPSCLIRKEP 95
Query: 146 VETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSV 205
+ +GEG E E R+ + +++ G + ++ +S+
Sbjct: 96 KDNNVSYEETGEGEYFEEEN------------RKNSNNANSSNSRNTGRGNKGGKERESL 143
Query: 206 NESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGS 265
N++ S+ + ++ + L + S++ +S L+ ++T + N
Sbjct: 144 NDNESSNEETEGRRSVKKTNTLNED--NNGNSIRKESNLSNDEETDEVVDT--HRINTED 199
Query: 266 YDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSE-VQSSSSLSKTRRTQRRRKE 324
+ +E K T + P ++RK + S ++ G G R +E + RR+ K+
Sbjct: 200 DEEDEEKPPTEKGTPKRKDRKSDSRSTSKGFGSGGRGRAEYTEDGDDDESNRRSVNFEKD 259
Query: 325 VSGEDAKMENE 335
+G K E E
Sbjct: 260 GNGRMVKTETE 270
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 39.3 bits (90), Expect = 0.016
Identities = 50/233 (21%), Positives = 98/233 (41%), Gaps = 18/233 (7%)
Query: 107 KQRVAELRRDVQLSDVSILSLQLQVKKLEDE---KAKENEEKVETEPDLAVSGEGRLPEN 163
KQ++ LR +++ + L Q+KK E++ K ++ E++ D S +
Sbjct: 1137 KQQLNSLRANLESLEKEHEDLAAQLKKYEEQIANKERQYNEEISQLNDEITSTQQENESI 1196
Query: 164 EKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEV 223
+K +++ V A++ E +N K A ++ + NE+N S +++K+ E
Sbjct: 1197 KKKNDELEGEVKAMKSTSEEQSNLKKSEIDALNLQIKELKKKNETNE-ASLLESIKSVES 1255
Query: 224 D-VKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSY--DNNEAKAVTCESVP 280
+ VK+ +Q + E + + + K E N Y E++ + E
Sbjct: 1256 ETVKI--------KELQDECNFKEKEVSELEDKLKASEDKNSKYLELQKESEKIKEELDA 1307
Query: 281 PSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAKME 333
+ E K++ + +T + SE+ S L KT +R+ E E K E
Sbjct: 1308 KTTELKIQLEKITNLSKAKEKSESEL---SRLKKTSSEERKNAEEQLEKLKNE 1357
Score = 38.5 bits (88), Expect = 0.028
Identities = 52/246 (21%), Positives = 111/246 (44%), Gaps = 32/246 (13%)
Query: 102 LDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLP 161
+++ K+ + L+ ++ + S ++ +KKLE+E +KE E +E ++ E +
Sbjct: 1457 IERDNKRDLESLKEQLRAAQESKAKVEEGLKKLEEESSKEKAE-LEKSKEMMKKLESTIE 1515
Query: 162 ENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTG 221
NE ++ + IR+ DE + K + EE ++ Q +E + SR +
Sbjct: 1516 SNET---ELKSSMETIRKSDEKLEQSKK----SAEEDIKNLQ--HEKSDLISR---INES 1563
Query: 222 EVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNG--SYDNNEAKAVTCESV 279
E D++ ++KS L ++G + ++V++E NN N + +S
Sbjct: 1564 EKDIE------------ELKSKLRIEAKSGSELETVKQELNNAQEKIRINAEENTVLKSK 1611
Query: 280 PPSEERKVEDDSVTRSEGGGTRESSEVQSS--SSLSKTRRTQRRRKEVSGEDAKMENEDV 337
ER+++D ++E +E E+ +S L + + +++ + S E+ + E
Sbjct: 1612 LEDIERELKD---KQAEIKSNQEEKELLTSRLKELEQELDSTQQKAQKSEEERRAEVRKF 1668
Query: 338 AVVKSE 343
V KS+
Sbjct: 1669 QVEKSQ 1674
>SERA_ENTHI (P21138) Serine-rich 25 kDa antigen protein (SHEHP)
(SREHP)
Length = 233
Score = 39.3 bits (90), Expect = 0.016
Identities = 38/202 (18%), Positives = 82/202 (39%), Gaps = 11/202 (5%)
Query: 110 VAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGD 169
+ +L ++V+ +++ + L+ + + E+A+E E+ +P+ + NE D
Sbjct: 17 ILDLDQEVKDTNIYGVFLKNEASPEKLEEAEEKEKSSSAKPESS--------SNEDNEDD 68
Query: 170 IDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGS 229
DE + + S+++ P E S D + S+ ++ +A + + D K +
Sbjct: 69 EDEKASSSDNSESSSSDKPDNKP---EASSSDKPEASSSDKPDNKPEASSSDKPDNKPEA 125
Query: 230 GPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVED 289
+ S ++PD S + ++ + N+ +A + S K E
Sbjct: 126 SSSDKPDNKPEASSSDKPDNKPEASSSDKPDNKPEASSTNKPEASSTNKPEASSTNKPEA 185
Query: 290 DSVTRSEGGGTRESSEVQSSSS 311
S + E T S++ SSS
Sbjct: 186 SSTNKPEASSTSNSNDKSGSSS 207
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 39.3 bits (90), Expect = 0.016
Identities = 39/256 (15%), Positives = 101/256 (39%), Gaps = 6/256 (2%)
Query: 84 QNGDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENE 143
++ D + ED + ++ RK++ A+ ++ + + + KK + ++E++
Sbjct: 9 EDSDGHVIEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDD 68
Query: 144 EKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQ 203
+ E P + + ++E + DE R+ +S D+ ++ +
Sbjct: 69 DDEEESPRKSSKKSRKRAKSESESDESDEEED--RKKSKSKKKVDQKKKEKSKKKRTTSS 126
Query: 204 SVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQ--VKSGLNEPDQTGRKRKSVEEES 261
S +E + + K + K S + VK +++ +KR EES
Sbjct: 127 SEDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEES 186
Query: 262 NNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRE--SSEVQSSSSLSKTRRTQ 319
+ + ++K + E + ED+ + +++ E +S + ++T+
Sbjct: 187 DEDEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTE 246
Query: 320 RRRKEVSGEDAKMENE 335
+R++ + + E+E
Sbjct: 247 KRKRSKTSSEESSESE 262
Score = 38.1 bits (87), Expect = 0.036
Identities = 53/259 (20%), Positives = 103/259 (39%), Gaps = 23/259 (8%)
Query: 93 AEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILSLQLQVKKLEDEKAKENEEKVETEPDL 152
+E D P K++ + +R S+ S S K E+E+ KE+ K + + L
Sbjct: 233 SESEDEAP-----EKKKTEKRKRSKTSSEESSES----EKSDEEEEEKESSPKPKKKKPL 283
Query: 153 AVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTNTDKLLPAAGEESDRDNQSVNESNSTG 212
AV +L +E++ E +P ++ T +D E ++D +S +E++
Sbjct: 284 AVK---KLSSDEESEESDVEVLPQKKKRGAVTLISDS-------EDEKDQKSESEASDVE 333
Query: 213 SRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNEPDQTGRKRKSVEEESNNGSYDNNEAK 272
+ K + + SG GS+ V + ++ +K+K + +S+ + +A+
Sbjct: 334 EKVSKKKAKKQESS-ESGSDSSEGSITVNRKSKKKEKPEKKKKGIIMDSSKLQKETIDAE 392
Query: 273 AVTCESVPPSEERKVEDDSVTRSEGGGTRESSEVQSSSSLSKTRRTQRRRKEVSGEDAK- 331
E E+++ E + + EG E SS K+ V E K
Sbjct: 393 RAEKERRKRLEKKQKEFNGIVLEEGEDLTEMLTGTSSQRKLKSVVLDPDSSTVDEESKKP 452
Query: 332 --MENEDVAVVKSEPLFGV 348
+ N V ++K G+
Sbjct: 453 VEVHNSLVRILKPHQAHGI 471
Score = 32.7 bits (73), Expect = 1.5
Identities = 51/275 (18%), Positives = 99/275 (35%), Gaps = 35/275 (12%)
Query: 73 RRFKDDVPPPQQN------GDVSAVTAEDSDHVPWLDKLRKQRVAELRRDVQLSDVSILS 126
+R ++ PPP++ S +D + P + ++ A+ + SD
Sbjct: 42 KREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSKKSRKRAKSESESDESDEEEDR 101
Query: 127 LQLQVKKLEDEKAKENEEKVETEPDLAVSGEGRLPENEKTGGDIDEPVPAIRRLDESTTN 186
+ + KK D+K KE +K T + D D ++ + +
Sbjct: 102 KKSKSKKKVDQKKKEKSKKKRTT---------------SSSEDEDSDEEREQKSKKKSKK 146
Query: 187 TDKLLPAAGEESDRDNQSVNESNSTGSRFDALKTGEVDVKLGSGPGHGSGSVQVKSGLNE 246
T K + E + + V +S K E VK + S + S +
Sbjct: 147 TKKQTSSESSEESEEERKVKKSK---------KNKEKSVKKRAETSEESDEDEKPS---K 194
Query: 247 PDQTGRKRKSVEEESNNGSYDNNEAKAVTCESVPPSEERKVEDDSVTRSEGGGTRESSEV 306
+ G K+K+ + ES + S D E K +S ++ +D + R+ S+
Sbjct: 195 KSKKGLKKKA-KSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKT 253
Query: 307 QSSSSLSKTRRTQRRRKEVSGEDAKMENEDVAVVK 341
S S S++ ++ +E + + +AV K
Sbjct: 254 SSEES-SESEKSDEEEEEKESSPKPKKKKPLAVKK 287
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.306 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,107,390
Number of Sequences: 164201
Number of extensions: 1941173
Number of successful extensions: 6141
Number of sequences better than 10.0: 318
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 298
Number of HSP's that attempted gapping in prelim test: 5718
Number of HSP's gapped (non-prelim): 629
length of query: 371
length of database: 59,974,054
effective HSP length: 112
effective length of query: 259
effective length of database: 41,583,542
effective search space: 10770137378
effective search space used: 10770137378
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146756.1


