

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
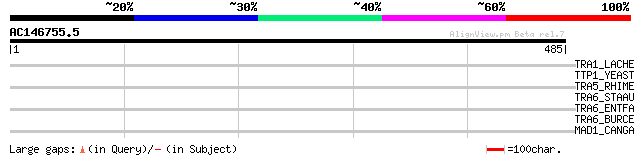
Query= AC146755.5 - phase: 0
(485 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TRA1_LACHE (P35880) Transposase for insertion sequence element I... 40 0.010
TTP1_YEAST (P38069) TTP1 protein (Mannan synthesis protein MNN2) 37 0.087
TRA5_RHIME (Q52873) Transposase for insertion sequence element I... 36 0.25
TRA6_STAAU (P19775) Transposase for insertion sequence element I... 33 1.6
TRA6_ENTFA (P59787) Transposase for insertion sequence element I... 33 1.6
TRA6_BURCE (P24575) Transposase for insertion sequence element I... 33 2.2
MAD1_CANGA (Q6FS63) Spindle assembly checkpoint component MAD1 (... 32 4.8
>TRA1_LACHE (P35880) Transposase for insertion sequence element
IS1201
Length = 369
Score = 40.4 bits (93), Expect = 0.010
Identities = 24/101 (23%), Positives = 51/101 (49%), Gaps = 5/101 (4%)
Query: 227 DATYKKIKYNT----PLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAMGGKL 282
DATY ++ T + I G+ + + I +E + + LL+ +++ G +
Sbjct: 163 DATYVPLRRETFEREAVYIAIGIKPNGHKEVIDYCIAPNENIEVWTELLQS-MKSRGLEQ 221
Query: 283 PVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIK 323
++DG + M+ A+ K +P+AH + C H++RN + ++
Sbjct: 222 VELFLSDGVVGMKTALAKTYPQAHFQRCLVHVMRNICAKVR 262
>TTP1_YEAST (P38069) TTP1 protein (Mannan synthesis protein MNN2)
Length = 597
Score = 37.4 bits (85), Expect = 0.087
Identities = 23/55 (41%), Positives = 29/55 (51%), Gaps = 1/55 (1%)
Query: 369 KRKMWAAAHFRGKFFAGFRTTSRCEGLHSEFGKYVSALTNLHDFFQQFFRWLNYM 423
K AAA+F G F RT + EG H E G + A+ HDF Q + R+LN M
Sbjct: 399 KETFIAAANFYGLSFYQVRTRTGVEGYHDEDGFHGVAMLQ-HDFVQDYGRYLNAM 452
>TRA5_RHIME (Q52873) Transposase for insertion sequence element
ISRM5
Length = 398
Score = 35.8 bits (81), Expect = 0.25
Identities = 27/108 (25%), Positives = 50/108 (46%), Gaps = 6/108 (5%)
Query: 224 LAFDATYKKIK-----YNTPLVIFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAM 278
L DA Y+K++ + ++I G++ + I + G E+ + L + ++
Sbjct: 161 LILDARYEKVREAGVVMSQAVLIAVGIDWDGRRQILSVEMAGRESRSAWKDFL-VRLKGR 219
Query: 279 GGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLH 326
G K V++D + AI +V PEA + C H +RNA ++ H
Sbjct: 220 GLKGVELVVSDDHAGLVAAIGEVIPEAAWQRCYVHFLRNALDHLPRKH 267
>TRA6_STAAU (P19775) Transposase for insertion sequence element
IS256 in transposon Tn4001
Length = 390
Score = 33.1 bits (74), Expect = 1.6
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 7/119 (5%)
Query: 224 LAFDATYKKIKYNTPLV-----IFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAM 278
L D Y K++ ++ I G+ I G I E+E+T+ + +++
Sbjct: 164 LMTDVLYIKVREENRVLSKSCHIAIGITKDGDREIIGFMIQSGESEETWTTFFE-YLKER 222
Query: 279 GGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDCLLG 337
G + VI+D + +AIRK F + C H +RN + I + S F++ + G
Sbjct: 223 GLQGTELVISDAHKGLVSAIRKSFTNVSWQRCQVHFLRNIFTTIPKKNSKS-FREAVKG 280
>TRA6_ENTFA (P59787) Transposase for insertion sequence element
IS256 in transposon Tn4001
Length = 390
Score = 33.1 bits (74), Expect = 1.6
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 7/119 (5%)
Query: 224 LAFDATYKKIKYNTPLV-----IFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAM 278
L D Y K++ ++ I G+ I G I E+E+T+ + +++
Sbjct: 164 LMTDVLYIKVREENRVLSKSCHIAIGITKDGDREIIGFMIQSGESEETWTTFFE-YLKER 222
Query: 279 GGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNATSNIKNLHFVSKFKDCLLG 337
G + VI+D + +AIRK F + C H +RN + I + S F++ + G
Sbjct: 223 GLQGTELVISDAHKGLVSAIRKSFTNVSWQRCQVHFLRNIFTTIPKKNSKS-FREAVKG 280
>TRA6_BURCE (P24575) Transposase for insertion sequence element
IS406
Length = 388
Score = 32.7 bits (73), Expect = 2.2
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 6/100 (6%)
Query: 224 LAFDATYKKIKYNTPLV-----IFSGVNHHNQSIIFGSAIIGDETEDTYVWLLKIFVEAM 278
L DA Y+K++ +V I G+ + + G + E E + L+ + A
Sbjct: 163 LFLDARYEKVRLEGRIVDCAVLIAVGIEASGKRRVLGCEVATSEAEINWRRFLESLL-AR 221
Query: 279 GGKLPVSVITDGDLSMRNAIRKVFPEAHHRLCAWHLIRNA 318
G K +I D ++ A R V P + C +HL +NA
Sbjct: 222 GLKGVTLIIADDHAGLKAARRAVLPSVPWQRCQFHLQQNA 261
>MAD1_CANGA (Q6FS63) Spindle assembly checkpoint component MAD1
(Mitotic arrest deficient protein 1)
Length = 657
Score = 31.6 bits (70), Expect = 4.8
Identities = 12/41 (29%), Positives = 23/41 (55%)
Query: 339 VDVDVFQRKWEELVTEFGLEENPWMLEMYQKRKMWAAAHFR 379
+DVD ++ K++ L+ ++ +E+ W E Y K A H +
Sbjct: 33 IDVDNWKYKYDNLLNQYEIEKVRWQKEQYDSEKSHEALHLQ 73
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,437,669
Number of Sequences: 164201
Number of extensions: 2240295
Number of successful extensions: 6273
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 6270
Number of HSP's gapped (non-prelim): 7
length of query: 485
length of database: 59,974,054
effective HSP length: 114
effective length of query: 371
effective length of database: 41,255,140
effective search space: 15305656940
effective search space used: 15305656940
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146755.5


