

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146755.11 + phase: 0
(246 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
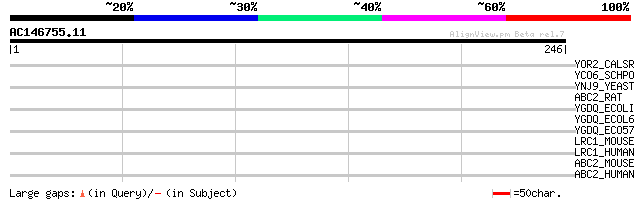
Score E
Sequences producing significant alignments: (bits) Value
YOR2_CALSR (P40980) Hypothetical 30.9 kDa protein in xylR 5'regi... 32 1.8
YCO6_SCHPO (O59797) Putative nucleosome assembly protein C364.06 31 2.4
YNJ9_YEAST (P50946) Hypothetical 27.2 kDa protein in POL1-RAS2 i... 30 4.1
ABC2_RAT (Q9ESR9) ATP-binding cassette, sub-family A, member 2 (... 30 7.0
YGDQ_ECOLI (P67127) Hypothetical UPF0053 protein ygdQ 29 9.2
YGDQ_ECOL6 (P67128) Hypothetical UPF0053 protein ygdQ 29 9.2
YGDQ_ECO57 (P67129) Hypothetical UPF0053 protein ygdQ 29 9.2
LRC1_MOUSE (P62046) Leucine-rich repeats and calponin homology d... 29 9.2
LRC1_HUMAN (Q9Y2L9) Leucine-rich repeats and calponin homology d... 29 9.2
ABC2_MOUSE (P41234) ATP-binding cassette, sub-family A, member 2... 29 9.2
ABC2_HUMAN (Q9BZC7) ATP-binding cassette, sub-family A, member 2... 29 9.2
>YOR2_CALSR (P40980) Hypothetical 30.9 kDa protein in xylR 5'region
(ORF2)
Length = 275
Score = 31.6 bits (70), Expect = 1.8
Identities = 16/76 (21%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query: 53 LAVVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKIL-KIYV 111
+ +++++S+ + F F L +++Y I+ +A P H + + T +IK+ ++
Sbjct: 80 VVLILIISSMAAFAFSRFKFALNNLLYSLIIAGMAIPIHVTLIPIYVLTNKIKLYDTVFA 139
Query: 112 FIGAIIGLYAPIAYIL 127
IG + L P++ +
Sbjct: 140 LIGPYVALSLPMSIFI 155
>YCO6_SCHPO (O59797) Putative nucleosome assembly protein C364.06
Length = 393
Score = 31.2 bits (69), Expect = 2.4
Identities = 33/124 (26%), Positives = 54/124 (42%), Gaps = 11/124 (8%)
Query: 18 EQEIEHKEEQDKSLNKSTSTPQRKTS-----FLSFRQLNCLAVVVVLSASGMVSPEDFGF 72
E+EI+ E D++ K ++ + K + A+ VLS S M++PED G
Sbjct: 130 EEEIKKGEAADENEKKEPTSSESKKQEGGDDTKGIPEFWLTAMKNVLSLSEMITPEDEGA 189
Query: 73 VLFSV-IYMCFISKVAFPSHPSKEQPPIFTQQIKILKIYVFIGAIIGLYAPIAYILHGIF 131
+ V I + ++ K F + P FT +I + K Y ++ P L+
Sbjct: 190 LSHLVDIRISYMEKPGFKLEFEFAENPFFTNKI-LTKTYYYMEE----SGPSNVFLYDHA 244
Query: 132 EGDK 135
EGDK
Sbjct: 245 EGDK 248
>YNJ9_YEAST (P50946) Hypothetical 27.2 kDa protein in POL1-RAS2
intergenic region
Length = 238
Score = 30.4 bits (67), Expect = 4.1
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 10/74 (13%)
Query: 11 DISLPKEEQEIEHKEEQDKS----LNKSTSTPQRKTSFLSFRQ------LNCLAVVVVLS 60
++S+P+EE E E +EE D +N+ T + + K Q LN V L
Sbjct: 22 NVSVPEEEVEDEDEEEDDDDDHIYINEETESGREKVLVSHAPQERIVPPLNFCPVERYLY 81
Query: 61 ASGMVSPEDFGFVL 74
SG SP +F F+L
Sbjct: 82 RSGQPSPVNFPFLL 95
>ABC2_RAT (Q9ESR9) ATP-binding cassette, sub-family A, member 2
(ATP-binding cassette transporter 2) (ATP-binding
cassette 2)
Length = 2434
Score = 29.6 bits (65), Expect = 7.0
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 13/89 (14%)
Query: 52 CLAVVVVLSASGMVSPEDFGFVL-FSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIY 110
C+ ++ V SP +F VL ++Y I+ + +P+ E P Y
Sbjct: 1855 CIIILFVFDLPAYTSPTNFPAVLSLFLLYGWSITPIMYPASFWFEVPS---------SAY 1905
Query: 111 VF---IGAIIGLYAPIAYILHGIFEGDKE 136
VF I IG+ A +A L +FE DK+
Sbjct: 1906 VFLIVINLFIGITATVATFLLQLFEHDKD 1934
>YGDQ_ECOLI (P67127) Hypothetical UPF0053 protein ygdQ
Length = 237
Score = 29.3 bits (64), Expect = 9.2
Identities = 21/106 (19%), Positives = 44/106 (40%), Gaps = 1/106 (0%)
Query: 55 VVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIYVFIG 114
VV L + G V+ + ++ +A+ + + IF+Q+I + + +G
Sbjct: 36 VVAKLPTAQRAHARRLGLAGAMVMRLALLASIAWVTRLTNPLFTIFSQEISARDLILLLG 95
Query: 115 AIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFS 160
+ ++ I H EG++EG+K Q+ + + FS
Sbjct: 96 GLFLIWKASKEI-HESIEGEEEGLKTRVSSFLGAIVQIMLLDIIFS 140
>YGDQ_ECOL6 (P67128) Hypothetical UPF0053 protein ygdQ
Length = 237
Score = 29.3 bits (64), Expect = 9.2
Identities = 21/106 (19%), Positives = 44/106 (40%), Gaps = 1/106 (0%)
Query: 55 VVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIYVFIG 114
VV L + G V+ + ++ +A+ + + IF+Q+I + + +G
Sbjct: 36 VVAKLPTAQRAHARRLGLAGAMVMRLALLASIAWVTRLTNPLFTIFSQEISARDLILLLG 95
Query: 115 AIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFS 160
+ ++ I H EG++EG+K Q+ + + FS
Sbjct: 96 GLFLIWKASKEI-HESIEGEEEGLKTRVSSFLGAIVQIMLLDIIFS 140
>YGDQ_ECO57 (P67129) Hypothetical UPF0053 protein ygdQ
Length = 237
Score = 29.3 bits (64), Expect = 9.2
Identities = 21/106 (19%), Positives = 44/106 (40%), Gaps = 1/106 (0%)
Query: 55 VVVVLSASGMVSPEDFGFVLFSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIYVFIG 114
VV L + G V+ + ++ +A+ + + IF+Q+I + + +G
Sbjct: 36 VVAKLPTAQRAHARRLGLAGAMVMRLALLASIAWVTRLTNPLFTIFSQEISARDLILLLG 95
Query: 115 AIIGLYAPIAYILHGIFEGDKEGIKAATPHVFLLASQVFMEGVAFS 160
+ ++ I H EG++EG+K Q+ + + FS
Sbjct: 96 GLFLIWKASKEI-HESIEGEEEGLKTRVSSFLGAIVQIMLLDIIFS 140
>LRC1_MOUSE (P62046) Leucine-rich repeats and calponin homology
domain containing protein 1 (Calponin homology domain
containing protein 1)
Length = 709
Score = 29.3 bits (64), Expect = 9.2
Identities = 15/39 (38%), Positives = 23/39 (58%)
Query: 13 SLPKEEQEIEHKEEQDKSLNKSTSTPQRKTSFLSFRQLN 51
SLP+E +++ E D S N+ T+ PQ+ S R+LN
Sbjct: 169 SLPEEIGQLKQLMELDVSCNEITALPQQIGQLKSLRELN 207
>LRC1_HUMAN (Q9Y2L9) Leucine-rich repeats and calponin homology
domain containing protein 1 (Calponin homology domain
containing protein 1) (Neuronal protein 81) (NP81)
Length = 728
Score = 29.3 bits (64), Expect = 9.2
Identities = 15/39 (38%), Positives = 23/39 (58%)
Query: 13 SLPKEEQEIEHKEEQDKSLNKSTSTPQRKTSFLSFRQLN 51
SLP+E +++ E D S N+ T+ PQ+ S R+LN
Sbjct: 179 SLPEEIGQLKQLMELDVSCNEITALPQQIGQLKSLRELN 217
>ABC2_MOUSE (P41234) ATP-binding cassette, sub-family A, member 2
(ATP-binding cassette transporter 2) (ATP-binding
cassette 2)
Length = 2434
Score = 29.3 bits (64), Expect = 9.2
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 13/89 (14%)
Query: 52 CLAVVVVLSASGMVSPEDFGFVL-FSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIY 110
C+ ++ V SP +F VL ++Y I+ + +P+ E P Y
Sbjct: 1855 CVIILFVFDLPAYTSPTNFPAVLSLFLLYGWSITPIMYPASFWFEVPS---------SAY 1905
Query: 111 VF---IGAIIGLYAPIAYILHGIFEGDKE 136
VF I IG+ A +A L +FE DK+
Sbjct: 1906 VFLIVINLFIGITATVATFLLQLFEHDKD 1934
>ABC2_HUMAN (Q9BZC7) ATP-binding cassette, sub-family A, member 2
(ATP-binding cassette transporter 2) (ATP-binding
cassette 2)
Length = 2436
Score = 29.3 bits (64), Expect = 9.2
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 13/89 (14%)
Query: 52 CLAVVVVLSASGMVSPEDFGFVL-FSVIYMCFISKVAFPSHPSKEQPPIFTQQIKILKIY 110
C+ ++ V SP +F VL ++Y I+ + +P+ E P Y
Sbjct: 1855 CVIILFVFDLPAYTSPTNFPAVLSLFLLYGWSITPIMYPASFWFEVPS---------SAY 1905
Query: 111 VF---IGAIIGLYAPIAYILHGIFEGDKE 136
VF I IG+ A +A L +FE DK+
Sbjct: 1906 VFLIVINLFIGITATVATFLLQLFEHDKD 1934
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,894,109
Number of Sequences: 164201
Number of extensions: 1111446
Number of successful extensions: 4292
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 4290
Number of HSP's gapped (non-prelim): 11
length of query: 246
length of database: 59,974,054
effective HSP length: 107
effective length of query: 139
effective length of database: 42,404,547
effective search space: 5894232033
effective search space used: 5894232033
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146755.11


