

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.14 - phase: 2 /pseudo
(1086 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
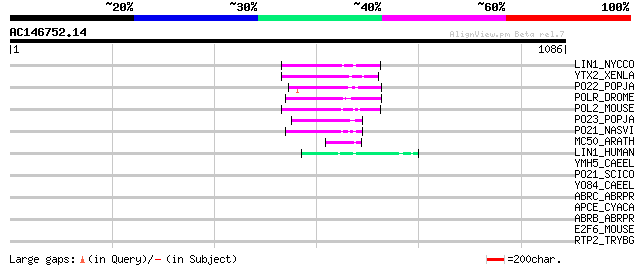
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 69 7e-11
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 67 3e-10
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 65 1e-09
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 58 1e-07
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 58 1e-07
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 58 1e-07
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 57 2e-07
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 53 5e-06
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 52 1e-05
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 43 0.004
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 39 0.076
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 35 1.4
ABRC_ABRPR (P28590) Abrin-c precursor [Contains: Abrin-c A chain... 34 2.4
APCE_CYACA (Q9TLS6) Phycobilisome linker polypeptide (Anchor pol... 33 3.2
ABRB_ABRPR (Q06077) Abrin-b precursor [Contains: Abrin-b A chain... 33 4.2
E2F6_MOUSE (O54917) Transcription factor E2F6 (E2F-6) (E2F-bindi... 32 7.1
RTP2_TRYBG (P15594) Retrotransposable element SLACS 132 kDa prot... 32 9.3
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 68.9 bits (167), Expect = 7e-11
Identities = 48/193 (24%), Positives = 87/193 (44%), Gaps = 7/193 (3%)
Query: 533 VGSVVFASQSAFIKGRQILDGILIANELVDDA-KRNNKELLMFKVDFEKAYDSVDWRYLD 591
+ ++ Q FI G Q I + ++ K NK+ ++ +D EKA+D++ ++
Sbjct: 554 IKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMI 613
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+ K+ + I + T ++++NG F G RQG PLSP LF + E
Sbjct: 614 RTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVME- 672
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
V+ + +G+E + FADD ++ + + + V+ + +V
Sbjct: 673 --VLAIAIREEKAIKGIHIGSE---EIKLSLFADDMIVYLENTRDSTTKLLEVIKEYSNV 727
Query: 712 SGLKVNFHKSMFF 724
SG K+N HKS+ F
Sbjct: 728 SGYKINTHKSVAF 740
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 66.6 bits (161), Expect = 3e-10
Identities = 52/190 (27%), Positives = 87/190 (45%), Gaps = 7/190 (3%)
Query: 532 VVGSVVFASQSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLD 591
V+ V+ QS + GR I D + + +L+ A+R L +D EKA+D VD +YL
Sbjct: 549 VLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEKAFDRVDHQYLI 608
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+ +F + ++ +A V +N T F RG+RQG PLS L+ LA E
Sbjct: 609 GTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEP 668
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
++ GL V E + V +ADD +L+ + ++ + ++
Sbjct: 669 FLCLLRK-RLTGL-----VLKEPDMRVVLSAYADDVILV-AQDLVDLERAQECQEVYAAA 721
Query: 712 SGLKVNFHKS 721
S ++N+ KS
Sbjct: 722 SSARINWSKS 731
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 65.1 bits (157), Expect = 1e-09
Identities = 57/188 (30%), Positives = 90/188 (47%), Gaps = 17/188 (9%)
Query: 546 KGRQILDGILIANELVD----DAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFPR 601
KG +DG L+ + L+D + K + +D KA+D+V + + ++
Sbjct: 108 KGYARIDGTLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDE 167
Query: 602 LWRSWIMECVTTATTSVLVN-GCPTDEFHFERGLRQGDPLSPFLF-LLAAEGLNVMMSTV 659
++I ++ +TT++ V G T + RG++QGDPLSPFLF + E L + ST
Sbjct: 168 GTSNYITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTP 227
Query: 660 GSNGLFAPYTVGAE-VPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNF 718
G G T+G E +PV L FADD LL+ + TV F + G+ +N
Sbjct: 228 GIGG-----TIGEEKIPV----LAFADDLLLLEDNDVLLPTTLATVANFFR-LRGMSLNA 277
Query: 719 HKSMFFGV 726
KS+ V
Sbjct: 278 KKSVSISV 285
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 58.2 bits (139), Expect = 1e-07
Identities = 43/186 (23%), Positives = 87/186 (46%), Gaps = 10/186 (5%)
Query: 541 QSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFP 600
Q F+ D I + ++ + ++ + + +D KA+DS+ + + + P
Sbjct: 446 QRGFLPTDGCADNATIVDLVLRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAP 505
Query: 601 RLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVG 660
+ + ++ TS+ +G ++EF RG++QGDPLSP LF L + L + T+
Sbjct: 506 KGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRL---LRTL- 561
Query: 661 SNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFHK 720
P +GA+V ++++ D L++ ++ ++ + L F + GLK+N K
Sbjct: 562 ------PSEIGAKVGNAITNAAAFADDLVLFAETRMGLQVLLDKTLDFLSIVGLKLNADK 615
Query: 721 SMFFGV 726
G+
Sbjct: 616 CFTVGI 621
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 58.2 bits (139), Expect = 1e-07
Identities = 48/193 (24%), Positives = 88/193 (44%), Gaps = 7/193 (3%)
Query: 533 VGSVVFASQSAFIKGRQILDGILIANELVDDA-KRNNKELLMFKVDFEKAYDSVDWRYLD 591
+ +++ Q FI G Q I + ++ K +K ++ +D EKA+D + ++
Sbjct: 581 IKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMIISLDAEKAFDKIQHPFMI 640
Query: 592 EVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEG 651
+V+ + + + I + ++ VNG + + G RQG PLSP+LF +
Sbjct: 641 KVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIV--- 697
Query: 652 LNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDV 711
L V+ + +G E V +S L ADD ++ + R + ++ F +V
Sbjct: 698 LEVLARAIRQQKEIKGIQIGKE-EVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEV 754
Query: 712 SGLKVNFHKSMFF 724
G K+N +KSM F
Sbjct: 755 VGYKINSNKSMAF 767
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 58.2 bits (139), Expect = 1e-07
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 7/140 (5%)
Query: 551 LDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFPRLWRSWIMEC 610
L I++ + + K + +D KA+D+V + M + +IM
Sbjct: 3 LANIIMLEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMST 62
Query: 611 VTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPYTV 670
+T A T+++V G T++ + G++QGDPLSP LF + + L ++ P
Sbjct: 63 ITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASMTP--- 119
Query: 671 GAEVPVSVSHLQFADDTLLI 690
++ L FADD LL+
Sbjct: 120 ----ACKIASLAFADDLLLL 135
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 57.4 bits (137), Expect = 2e-07
Identities = 42/150 (28%), Positives = 74/150 (49%), Gaps = 9/150 (6%)
Query: 541 QSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFP 600
Q AFI + + + + ++ +A+ K L + +D +KA+DSV+ R + + + + P
Sbjct: 420 QRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLP 479
Query: 601 RLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVG 660
R++IM + T + V RG+RQGDPLSP LF ++ ++ +
Sbjct: 480 LEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCV---MDAVLRRLP 536
Query: 661 SNGLFAPYTVGAEVPVSVSHLQFADDTLLI 690
N + +GAE + L FADD +L+
Sbjct: 537 EN---TGFLMGAE---KIGALVFADDLVLL 560
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 52.8 bits (125), Expect = 5e-06
Identities = 29/69 (42%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query: 619 LVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPYTVGAEVPVSV 678
++NG P RGLRQGDPLSP+LF+L E L+ + G V P +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSP-RI 71
Query: 679 SHLQFADDT 687
+HL FADDT
Sbjct: 72 NHLLFADDT 80
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 51.6 bits (122), Expect = 1e-05
Identities = 51/228 (22%), Positives = 90/228 (39%), Gaps = 12/228 (5%)
Query: 572 LMFKVDFEKAYDSVDWRYLDEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFE 631
++ +D EKA+D + ++ + + K+ + I T ++++NG + +
Sbjct: 594 MIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLK 653
Query: 632 RGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIG 691
G RQG PLSP LL L V+ + +G E V FADD ++
Sbjct: 654 TGTRQGCPLSP---LLPNIVLEVLARAIRQEKEIKGIQLGKE---EVKLSLFADDMIVYL 707
Query: 692 VKSWANVRAMKTVLLLFEDVSGLKVNFHKSMFFGVNINESWLHEAAVVMHCRHGRIPFIY 751
+ + + ++ F VSG K+N KS F N + + Y
Sbjct: 708 ENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKY 767
Query: 752 LGLPIGGDPRKLHFWHPLVERIRRRLSGVLEDRESLWNVVLCAKYGEV 799
LG+ + D + L E + L+ + ED W + C+ G +
Sbjct: 768 LGIQLTRDVKDL-----FKENYKPLLNEIKEDTNK-WKNIPCSWVGRI 809
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 43.1 bits (100), Expect = 0.004
Identities = 30/108 (27%), Positives = 47/108 (42%), Gaps = 1/108 (0%)
Query: 541 QSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFP 600
Q F+ R ++ + L +N K L + DF KA+D V L + +A
Sbjct: 718 QHGFLNFRSCPSSLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLD 777
Query: 601 RLWRSWIMECVTTATTSVLVNG-CPTDEFHFERGLRQGDPLSPFLFLL 647
+L SW E + T SV +N ++ + G+ QG P LF+L
Sbjct: 778 KLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 38.9 bits (89), Expect = 0.076
Identities = 39/152 (25%), Positives = 69/152 (44%), Gaps = 10/152 (6%)
Query: 541 QSAFIKGRQILDGILIANELVDDAKRNNKELLMFKVDFEKAYDSVDWRYLDEVMAKMNFP 600
Q+A++ + + + ++ +AKR KEL + +D KA++SV L + + + P
Sbjct: 261 QTAYLPIDGVCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCP 320
Query: 601 RLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLRQGDPLSPFLFLLAAEGLNVMMSTVG 660
+I + T + G + G+ QGDPLS LF LA E ++ G
Sbjct: 321 PGVVDYIADMYNNVITEMQFEG-KCELASILAGVYQGDPLSGPLFTLAYEKALRALNNEG 379
Query: 661 SNGLFAPYTVGAEVPVSVSHLQFADDTLLIGV 692
+ A+V V+ S ++DD LL+ +
Sbjct: 380 RFDI-------ADVRVNAS--AYSDDGLLLAM 402
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 34.7 bits (78), Expect = 1.4
Identities = 30/114 (26%), Positives = 46/114 (40%), Gaps = 18/114 (15%)
Query: 576 VDFEKAYDSVDWRYLDEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGLR 635
+DF KA+D V L + + + + W+ +T + V V ++ G+
Sbjct: 23 LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRSFKVKVGNTLSEPKKTVCGVP 82
Query: 636 QGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLL 689
QG +SP LF G+F + A +PV V QFADD L
Sbjct: 83 QGSVISPVLF-----------------GIFV-NEISANLPVGVYCKQFADDIKL 118
>ABRC_ABRPR (P28590) Abrin-c precursor [Contains: Abrin-c A chain
(EC 3.2.2.22) (rRNA N-glycosidase); Abrin-c B chain]
Length = 562
Score = 33.9 bits (76), Expect = 2.4
Identities = 40/149 (26%), Positives = 64/149 (42%), Gaps = 28/149 (18%)
Query: 625 TDEFHFERGLRQGDPLSPFL--------FLLAAEGLNVMMSTVGSN---GLFAPYTVGAE 673
T+E+ +G R G+ SPF+ + A+G NV ++ +N +A YT G+
Sbjct: 422 TNEYLMRQGWRTGNNTSPFVTSISGYSDLCMQAQGSNVWLADCDNNKKEQQWALYTDGSI 481
Query: 674 VPV--------SVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFHKSMFFG 725
V S H Q + L+ WA+ R LF++ G N H M
Sbjct: 482 RSVQNTNNCLTSKDHKQGSPIVLMACSNGWASQR------WLFKN-DGSIYNLHDDMVMD 534
Query: 726 VNINESWLHEAAVVMHCRHGRIPFIYLGL 754
V ++ L E +++H HG+ I+L L
Sbjct: 535 VKRSDPSLKE--IILHPYHGKPNQIWLTL 561
>APCE_CYACA (Q9TLS6) Phycobilisome linker polypeptide (Anchor
polypeptide) (PBS-anchor protein)
Length = 870
Score = 33.5 bits (75), Expect = 3.2
Identities = 47/193 (24%), Positives = 78/193 (40%), Gaps = 24/193 (12%)
Query: 632 RGLRQGDPLSPFLFLLAAEGLNVMMSTVGSNGLFAPYTVGAEVPVSVSHLQFADDTLLIG 691
RGL + +L++ GL+ ++ ++ +N +A Y VP + Q A + G
Sbjct: 347 RGLSSLQEFRKYFAILSSNGLDALIDSIINNSEYAEYFGEETVPYIRGYGQEAQECRNWG 406
Query: 692 -----VKSWANVRAMKTVLLLFEDVSGL--------KVNFHKSMFFGVNINESWLHE-AA 737
K A R + + LF D + L K+N ++ FG S+++E +
Sbjct: 407 SQFALFKYSAPFRTIPQFITLFADYTQLPPSQHCYGKLNDPLNIQFGAIFKNSYVNEQSR 466
Query: 738 VVMHCRHGRIPFIYLGLPIG---GDPRKLHFWHPLVERIRRRLSGVLEDRESLWNVVLCA 794
V+ R R +Y G I G P L E+ +S +E+ N +L A
Sbjct: 467 PVLFPRGSRRILVYKGAGIFNQLGSPNAL-------EKPPSNVSIAKWSKETDLNFILNA 519
Query: 795 KYGEVGGRVQFSE 807
Y V GR + E
Sbjct: 520 AYLRVFGRYVYEE 532
>ABRB_ABRPR (Q06077) Abrin-b precursor [Contains: Abrin-b A chain
(EC 3.2.2.22) (rRNA N-glycosidase); Abrin-b B chain]
Length = 527
Score = 33.1 bits (74), Expect = 4.2
Identities = 40/149 (26%), Positives = 63/149 (41%), Gaps = 28/149 (18%)
Query: 625 TDEFHFERGLRQGDPLSPFL--------FLLAAEGLNVMMSTVGSN---GLFAPYTVGAE 673
T+E+ +G R G+ SPF+ + A+G NV ++ +N +A YT G+
Sbjct: 387 TNEYLMRQGWRTGNNTSPFVTSISGYSDLCMQAQGSNVWLAYCDNNKKEQQWALYTDGSI 446
Query: 674 VPV--------SVSHLQFADDTLLIGVKSWANVRAMKTVLLLFEDVSGLKVNFHKSMFFG 725
V S H Q + L+ WA+ R LF + G N H M
Sbjct: 447 RSVQNTNNCLTSKDHKQGSPIVLMACSNGWASQR------WLFRN-DGSIYNLHDDMVMD 499
Query: 726 VNINESWLHEAAVVMHCRHGRIPFIYLGL 754
V ++ L E +++H HG+ I+L L
Sbjct: 500 VKRSDPSLKE--IILHPYHGKPNQIWLTL 526
>E2F6_MOUSE (O54917) Transcription factor E2F6 (E2F-6) (E2F-binding
site modulating activity protein) (EMA)
Length = 272
Score = 32.3 bits (72), Expect = 7.1
Identities = 15/38 (39%), Positives = 26/38 (67%)
Query: 976 LFLNRLATKDNLRKRNVLEATNVSSGALCGKEEERDHL 1013
L LN++ATK ++RKR V + TNV G +++ ++H+
Sbjct: 87 LDLNKVATKLSVRKRRVYDITNVLDGIELVEKKSKNHI 124
>RTP2_TRYBG (P15594) Retrotransposable element SLACS 132 kDa protein
(ORF2)
Length = 1182
Score = 32.0 bits (71), Expect = 9.3
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 1/71 (1%)
Query: 576 VDFEKAYDSVDWR-YLDEVMAKMNFPRLWRSWIMECVTTATTSVLVNGCPTDEFHFERGL 634
+D AY+++ R L+ V + LWR + TT NG + RG+
Sbjct: 646 LDGRNAYNAISRRAILEAVYGDSTWSPLWRLVSLLLGTTGEVGFYENGKLCHTWESTRGV 705
Query: 635 RQGDPLSPFLF 645
RQG L P LF
Sbjct: 706 RQGMVLGPLLF 716
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.347 0.155 0.542
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 115,817,284
Number of Sequences: 164201
Number of extensions: 4567859
Number of successful extensions: 17516
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 17493
Number of HSP's gapped (non-prelim): 21
length of query: 1086
length of database: 59,974,054
effective HSP length: 121
effective length of query: 965
effective length of database: 40,105,733
effective search space: 38702032345
effective search space used: 38702032345
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146752.14


