

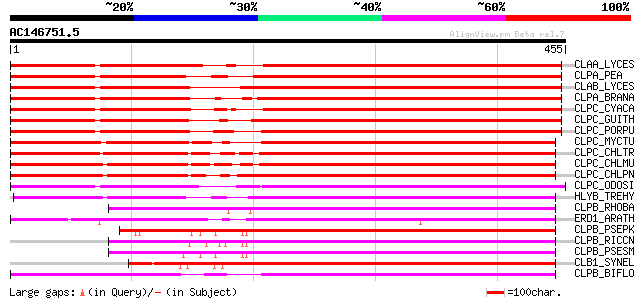
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146751.5 + phase: 0 /pseudo
(455 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CLAA_LYCES (P31541) ATP-dependent Clp protease ATP-binding subun... 412 e-115
CLPA_PEA (P35100) ATP-dependent Clp protease ATP-binding subunit... 409 e-114
CLAB_LYCES (P31542) ATP-dependent Clp protease ATP-binding subun... 409 e-113
CLPA_BRANA (P46523) ATP-dependent Clp protease ATP-binding subun... 406 e-113
CLPC_CYACA (Q9TM05) ATP-dependent Clp protease ATP-binding subun... 377 e-104
CLPC_GUITH (O78410) ATP-dependent Clp protease ATP-binding subun... 376 e-104
CLPC_PORPU (P51332) ATP-dependent Clp protease ATP-binding subun... 364 e-100
CLPC_MYCTU (O06286) Probable ATP-dependent Clp protease ATP-bind... 349 1e-95
CLPC_CHLTR (O84288) Probable ATP-dependent Clp protease ATP-bind... 347 5e-95
CLPC_CHLMU (Q9PKA8) Probable ATP-dependent Clp protease ATP-bind... 345 2e-94
CLPC_CHLPN (Q9Z8A6) Probable ATP-dependent Clp protease ATP-bind... 341 2e-93
CLPC_ODOSI (P49574) ATP-dependent Clp protease ATP-binding subun... 315 2e-85
HLYB_TREHY (Q54316) Hemolysin B 299 9e-81
CLPB_RHOBA (Q7UM33) Chaperone clpB 277 5e-74
ERD1_ARATH (P42762) ERD1 protein, chloroplast precursor 263 9e-70
CLPB_PSEPK (Q88Q71) Chaperone clpB 260 6e-69
CLPB_RICCN (Q92JK8) Chaperone clpB 259 8e-69
CLPB_PSESM (Q889C2) Chaperone clpB 256 1e-67
CLB1_SYNEL (Q8DJ40) Chaperone clpB 1 256 1e-67
CLPB_BIFLO (Q8G4X4) Chaperone clpB 255 1e-67
>CLAA_LYCES (P31541) ATP-dependent Clp protease ATP-binding subunit
clpA homolog CD4A, chloroplast precursor
Length = 926
Score = 412 bits (1060), Expect = e-115
Identities = 229/453 (50%), Positives = 299/453 (65%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 93 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 152
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 153 VEVEKIIGRG---SGFIAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 209
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD IR QVIR + E+ V G ++
Sbjct: 210 GVAARVLENLGADPTNIRTQVIRMVGESSEAVGASVGGG------------------TSG 251
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+K P L E +GTNLTKLA+EGKL P VGR+ Q+ERV QI+
Sbjct: 252 LKMPTL---------------------EEYGTNLTKLAEEGKLDPVVGRQAQIERVTQIL 290
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI +G VPE ++GKKV+ LD+ + +G
Sbjct: 291 GRRTKNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRGE 350
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ L++EI+ +ILF+ EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 351 FEERLKKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 410
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPVKV EPSV+ETI+ILKGLR YE H+KLHYTDEA+ AAA
Sbjct: 411 LDEYRKHIEKDPALERRFQPVKVPEPSVDETIQILKGLRERYEIHHKLHYTDEAIEAAAK 470
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 471 LSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQL 503
>CLPA_PEA (P35100) ATP-dependent Clp protease ATP-binding subunit
clpA homolog, chloroplast precursor
Length = 922
Score = 409 bits (1052), Expect = e-114
Identities = 226/453 (49%), Positives = 301/453 (65%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 91 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 150
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ S + A+ LG++ + + HLLLGLL+
Sbjct: 151 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSQEEARQLGHNYIGSEHLLLGLLREGE 207
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD IR QVIR + G+ ++VTA +
Sbjct: 208 GVAARVLENLGADPTNIRTQVIRMV-------------------GESADSVTATVGSGS- 247
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+N + LE +GTNLTKLA+EGKL P VGR+ Q+ERV QI+
Sbjct: 248 -------------------SNNKTPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERVTQIL 288
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI +G VPE ++GKKV+ LD+ + +G
Sbjct: 289 GRRTKNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRGE 348
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ L++EI+ ++ILF+ EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 349 FEERLKKLMEEIKQSDDIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 408
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTDEAL+AAA
Sbjct: 409 LDEYRKHIEKDPDLERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDEALIAAAQ 468
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPDKAIDL+DEAGS V+L HA++
Sbjct: 469 LSYQYISDRFLPDKAIDLVDEAGSRVRLQHAQL 501
>CLAB_LYCES (P31542) ATP-dependent Clp protease ATP-binding subunit
clpA homolog CD4B, chloroplast precursor
Length = 923
Score = 409 bits (1050), Expect = e-113
Identities = 229/454 (50%), Positives = 299/454 (65%), Gaps = 45/454 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 91 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 150
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 151 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 207
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD + IR QVIR + E+
Sbjct: 208 GVAARVLENLGADPSNIRTQVIRMVGES-------------------------------- 235
Query: 181 IKFPVLTQNNNLGARKDDNANKQKS-ALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQI 239
N +GA + QK LE +GTNLTKLA+EGKL P VGR+ Q+ERV QI
Sbjct: 236 --------NEAVGASVGGGTSGQKMPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERVTQI 287
Query: 240 ICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQG 299
+ RR KNNPCL+GEPGVGKT+I +GLAQRI +G VPE ++GKKV+ LD+ + +G
Sbjct: 288 LGRRTKNNPCLIGEPGVGKTAIAEGLAQRIANGDVPETIEGKKVITLDMGLLVAGTKYRG 347
Query: 300 SSEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFAT 358
EER++ L++EI+ +ILF+ EVH + A A GA A ILK AL RG +QCI AT
Sbjct: 348 EFEERLKKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGAT 407
Query: 359 TVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAA 418
T++E+R H+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTDE LVAAA
Sbjct: 408 TLDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDEDLVAAA 467
Query: 419 NLSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPDKAIDLIDEAGS V+L HA++
Sbjct: 468 QLSYQYISDRFLPDKAIDLIDEAGSRVRLRHAQL 501
>CLPA_BRANA (P46523) ATP-dependent Clp protease ATP-binding subunit
clpA homolog, chloroplast precursor (Fragment)
Length = 874
Score = 406 bits (1043), Expect = e-113
Identities = 230/453 (50%), Positives = 298/453 (65%), Gaps = 44/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS GI+ +R
Sbjct: 41 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDAR 100
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + + HLLLGLL+
Sbjct: 101 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGE 157
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N GAD + IR QVIR + EN VTA
Sbjct: 158 GVAARVLENLGADPSNIRTQVIRMVGEN--------------------NEVTA------- 190
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
N+G N + LE +GTNLTKLA+EGKL P VGR Q+ERV+QI+
Sbjct: 191 ----------NVGGGSGTN---KMPTLEEYGTNLTKLAEEGKLDPVVGRHPQIERVVQIL 237
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNPCL+GEPGVGKT+I +GLAQRI SG V E +GKKV+ LD+ +G
Sbjct: 238 GRRTKNNPCLIGEPGVGKTAIAEGLAQRIASGVVRETSEGKKVITLDMGLLAAGTKYRGE 297
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ L++EI+ +ILF+ EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 298 FEERVKKLMEEIKQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 357
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQPVKV EP+V+ETI+ILKGLR YE H+KL YTDE+LVAAA
Sbjct: 358 LDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQ 417
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+S+RFLPD+AIDL+DEAGS V+L HA+V
Sbjct: 418 LSYQYISDRFLPDRAIDLMDEAGSRVRLRHAQV 450
>CLPC_CYACA (Q9TM05) ATP-dependent Clp protease ATP-binding subunit
clpA homolog
Length = 854
Score = 377 bits (969), Expect = e-104
Identities = 216/453 (47%), Positives = 290/453 (63%), Gaps = 46/453 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLGILG+ GLAAK LKS GI +R
Sbjct: 32 MFERFTEKAVKVIMLAQEEARRLGHNFVGTEQILLGILGEGTGLAAKALKSMGITLKDAR 91
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK +L+ +++ ++ L ++ V T HLLLGL++
Sbjct: 92 IEVEKIIGRG---SGFVAIEIPFTPRAKKILELAIEESRILTHNYVGTEHLLLGLIKEGE 148
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N G D+ K+R +IR I E E +V A S +S
Sbjct: 149 GVAARVLENLGVDLPKLRSNIIRMIGETS------------------EVSVGATSGRS-- 188
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
K P L E FGTNLT++A EGKL P VGR +++ERV+QI+
Sbjct: 189 -KVPTL---------------------EEFGTNLTQMAVEGKLDPVVGRAKEIERVVQIL 226
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI++ VP+ L+ KKV+ LDV+ + +G
Sbjct: 227 GRRTKNNPVLIGEPGVGKTAIAEGLAQRIINNEVPDTLEDKKVITLDVSLLVAGTKYRGE 286
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI + NVIL + EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 287 FEERLKKIMDEIRMADNVILVIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATT 346
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R H+E D L+R FQPV V EP+VEETIEIL+GLR YE+H++L +D A+VAAA
Sbjct: 347 LEEYRKHIEKDAALERRFQPVMVEEPTVEETIEILRGLRDRYEAHHRLKISDSAIVAAAK 406
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
LS QY+++RFLPDKAIDL+DEA S V+L + K+
Sbjct: 407 LSDQYIADRFLPDKAIDLVDEASSRVRLMNYKL 439
>CLPC_GUITH (O78410) ATP-dependent Clp protease ATP-binding subunit
clpA homolog
Length = 819
Score = 376 bits (966), Expect = e-104
Identities = 214/453 (47%), Positives = 289/453 (63%), Gaps = 46/453 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AAK+LKS G++ +R
Sbjct: 1 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGVNLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLLLGL++
Sbjct: 61 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N D+ K+R QVIR + + TAE S +N
Sbjct: 118 GVAARVLENLALDLTKVRTQVIRLLGD------------------------TAEVSATN- 152
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ LE FG+NLT+ A EGKL P +GR++++ERVIQI+
Sbjct: 153 -----------------GQTKGKTPTLEEFGSNLTQKAAEGKLDPVIGRQKEIERVIQIL 195
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI + VP+ L+ K+VV LD+ + +G
Sbjct: 196 GRRTKNNPILIGEPGVGKTAIAEGLAQRINNRDVPDILEDKRVVTLDIGLLVAGTKYRGE 255
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ +I EI + NVIL + EVH + A A GA A ILK AL RG +QCI ATT
Sbjct: 256 FEERLKKIIDEIRVANNVILVIDEVHTLIGAGAAEGAIDAANILKPALARGEMQCIGATT 315
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R H+E D+ L+R FQPV V EPSVEETIEIL GLR YE H+KL +DEAL AAA
Sbjct: 316 LEEYRKHIEKDSALERRFQPVMVGEPSVEETIEILYGLRDRYEKHHKLVISDEALSAAAK 375
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
+ QY+++RFLPDKAIDLIDEAGS V+L ++++
Sbjct: 376 FADQYIADRFLPDKAIDLIDEAGSRVRLMNSQL 408
>CLPC_PORPU (P51332) ATP-dependent Clp protease ATP-binding subunit
clpA homolog
Length = 821
Score = 364 bits (935), Expect = e-100
Identities = 208/453 (45%), Positives = 286/453 (62%), Gaps = 43/453 (9%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM AQ+EAR +GH ++ TE ILLG++G+ G+AA++LKS ++ +R
Sbjct: 1 MFERFTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLVGEGTGIAAQVLKSMNVNLKDAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V K++GRG SGF +I FT AK VL+ SL+ A+ LG++ + T HLL+GL++
Sbjct: 61 VEVEKIIGRG---SGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGTEHLLMGLVREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G ++++N DV+ IR +VI+ + EN E NV+ ++ +
Sbjct: 118 GVAARVLENLAVDVSSIRAEVIQMLGEN------------------AEANVSGSNATQAR 159
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
K P LE FG+NLT++A EG L P VGR++++ERVIQI+
Sbjct: 160 SKTP---------------------TLEEFGSNLTQMAIEGGLDPVVGRQKEIERVIQIL 198
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT+I +GLAQRI + VP L+ K V+ LDV + +G
Sbjct: 199 GRRTKNNPVLIGEPGVGKTAIAEGLAQRIANRDVPSILEDKLVITLDVGLLVAGTKYRGE 258
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++ EI+ NVIL + EVH + A A GA A +LK AL RG +QCI ATT
Sbjct: 259 FEERLKRIMDEIKSADNVILVIDEVHTLIGAGAAEGAIDAANLLKPALARGELQCIGATT 318
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
+ E+R H+E D L+R F PV V EPSVEETIEIL GLR YE H++L +D AL AAA
Sbjct: 319 LEEYRKHIEKDPALERRFHPVVVGEPSVEETIEILFGLRDRYEKHHQLTMSDGALAAAAK 378
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKV 452
+ QY+S+RFLPDKAIDLIDEAGS V+L ++++
Sbjct: 379 YANQYISDRFLPDKAIDLIDEAGSRVRLLNSQL 411
>CLPC_MYCTU (O06286) Probable ATP-dependent Clp protease ATP-binding
subunit
Length = 848
Score = 349 bits (895), Expect = 1e-95
Identities = 194/448 (43%), Positives = 281/448 (62%), Gaps = 37/448 (8%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T+ A++V++ AQ+EAR + H YI TEHILLG++ + G+AAK L+S GI R
Sbjct: 1 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLESLGISLEGVR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
QV +++G+G I FT AK VL+ SL+ A LG++ + T H+LLGL++
Sbjct: 61 SQVEEIIGQGQQAPS---GHIPFTPRAKKVLELSLREALQLGHNYIGTEHILLGLIREGE 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q++ GA++ ++R+QVI+ + +Q AE GG+ S S
Sbjct: 118 GVAAQVLVKLGAELTRVRQQVIQLLSG--YQGKEAAEAGTGGRGGE-------SGSPSTS 168
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ L+ FG NLT A EGKL P +GRE+++ERV+Q++
Sbjct: 169 L------------------------VLDQFGRNLTAAAMEGKLDPVIGREKEIERVMQVL 204
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++GLAQ I+ G VPE LK K++ LD+ + +G
Sbjct: 205 SRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTLDLGSLVAGSRYRGD 264
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EER++ ++KEI G++ILF+ E+H + A A GA A ILK L RG +Q I ATT
Sbjct: 265 FEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEGAIDAASILKPKLARGELQTIGATT 324
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R ++E D L+R FQPV+V EP+VE TIEILKGLR YE+H+++ TD A+VAAA
Sbjct: 325 LDEYRKYIEKDAALERRFQPVQVGEPTVEHTIEILKGLRDRYEAHHRVSITDAAMVAAAT 384
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
L+ +Y+++RFLPDKAIDLIDEAG+ +++
Sbjct: 385 LADRYINDRFLPDKAIDLIDEAGARMRI 412
>CLPC_CHLTR (O84288) Probable ATP-dependent Clp protease ATP-binding
subunit
Length = 854
Score = 347 bits (889), Expect = 5e-95
Identities = 194/448 (43%), Positives = 284/448 (63%), Gaps = 19/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRTLGVDFDTAK 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V +L+G G C D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 HEVERLIGYGPEIQ--VCGDPALTGRVKKSFESANEEAALLEHNYVGTEHLLLGILNQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D +IR+++++++E + LP S SS S++
Sbjct: 119 GVALQVLENLHVDPKEIRKEILKELE--TFNLQLPPSSSITPRN--------TNSSSSSK 168
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
P+ + LG K + SAL+ +G +LT++ +E +L P +GR +VER+I I+
Sbjct: 169 SSSPL--GGHTLGGDKPEKL----SALKAYGYDLTEMFKESRLDPVIGRSAEVERLILIL 222
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP LVGE GVGKT+I++GLAQ+I+SG VPE L+ K+++ LD+A + +G
Sbjct: 223 CRRRKNNPVLVGEAGVGKTAIVEGLAQKIVSGEVPEALRKKRLITLDLALMIAGTKYRGQ 282
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA ++ILK AL RG IQCI ATT
Sbjct: 283 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASHILKPALARGEIQCIGATT 342
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ET+EIL+GL+ YE H+ + TDEALVAAA
Sbjct: 343 LDEYRKHIEKDAALERRFQKIVVQPPSVDETVEILRGLKKKYEEHHNVFITDEALVAAAK 402
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 403 LSDQYVHGRFLPDKAIDLLDEAGARVRV 430
>CLPC_CHLMU (Q9PKA8) Probable ATP-dependent Clp protease ATP-binding
subunit
Length = 870
Score = 345 bits (884), Expect = 2e-94
Identities = 191/448 (42%), Positives = 287/448 (63%), Gaps = 18/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ G+DF+ ++
Sbjct: 17 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRTLGVDFDTAK 76
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
+V +L+G G + D T K + + + A L ++ V T HLLLG+L ++
Sbjct: 77 NEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASILEHNYVGTEHLLLGILNQAD 134
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G Q+++N D +IR+++++++E + LP S + ++ SSKS+
Sbjct: 135 GVALQVLENLHIDPKEIRKEILKELE--TFNLQLPPSSSITPRN---TNSSSSSSSKSSS 189
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ ++LG K + SAL+ +G +LT++ +E +L P +GR +VER+I I+
Sbjct: 190 LG------GHSLGGDKPEKL----SALKAYGYDLTEMFKEARLDPVIGRSAEVERLILIL 239
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+SG VPE L+ K+++ LD+A + +G
Sbjct: 240 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIVSGEVPEALRKKRLITLDLALMIAGTKYRGQ 299
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA ++ILK AL RG IQCI ATT
Sbjct: 300 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASHILKPALARGEIQCIGATT 359
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ET+EIL+GL+ YE H+ + TDEALVAAA
Sbjct: 360 LDEYRKHIEKDAALERRFQKIVVQPPSVDETVEILRGLKKKYEEHHNVFITDEALVAAAK 419
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 420 LSDQYVHGRFLPDKAIDLLDEAGARVRV 447
>CLPC_CHLPN (Q9Z8A6) Probable ATP-dependent Clp protease ATP-binding
subunit
Length = 845
Score = 341 bits (875), Expect = 2e-93
Identities = 189/448 (42%), Positives = 283/448 (62%), Gaps = 21/448 (4%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E T AK+VI A+KEA+ + H Y+ TEHILLG+L G+A +L++ GIDF+ +R
Sbjct: 1 MFEKFTNRAKQVIKLAKKEAQRLNHNYLGTEHILLGLLKLGQGVAVNVLRNLGIDFDTAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++V +L+G G + D T K + + + A L ++ V T HLLLG+L S+
Sbjct: 61 QEVERLIGYGPEIQVYG--DPALTGRVKKSFESANEEASLLEHNYVGTEHLLLGILHQSD 118
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
Q+++N D ++R+++++++E + LP S + + SS+SN
Sbjct: 119 SVALQVLENLHIDPREVRKEILKELE--TFNLQLPPSSSSS-----------SSSSRSNP 165
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
+ + LG + N++ SAL+ +G +LT++ +E KL P +GR +VER+I I+
Sbjct: 166 S-----SSKSPLGHSLGSDKNEKLSALKAYGYDLTEMVRESKLDPVIGRSSEVERLILIL 220
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
CRR KNNP L+GE GVGKT+I++GLAQ+I+ VP+ L+ K+++ LD+A + +G
Sbjct: 221 CRRRKNNPVLIGEAGVGKTAIVEGLAQKIILNEVPDALRKKRLITLDLALMIAGTKYRGQ 280
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ ++ E+ GN++LF+ E+H I A A GA + ILK AL RG IQCI ATT
Sbjct: 281 FEERIKAVMDEVRKHGNILLFIDELHTIVGAGAAEGAIDASNILKPALARGEIQCIGATT 340
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R H+E D L+R FQ + V PSV+ETIEIL+GL+ YE H+ + T+EAL AAA
Sbjct: 341 IDEYRKHIEKDAALERRFQKIVVHPPSVDETIEILRGLKKKYEEHHNVFITEEALKAAAT 400
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS QYV RFLPDKAIDL+DEAG+ V++
Sbjct: 401 LSDQYVHGRFLPDKAIDLLDEAGARVRV 428
>CLPC_ODOSI (P49574) ATP-dependent Clp protease ATP-binding subunit
clpA homolog
Length = 885
Score = 315 bits (806), Expect = 2e-85
Identities = 175/456 (38%), Positives = 270/456 (58%), Gaps = 35/456 (7%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M E TE A KVIM +Q+EAR +GH ++ TE +LLGI+G +G+ A+ LK + +R
Sbjct: 1 MFEKFTEGAIKVIMLSQEEARRMGHNFVGTEQLLLGIIGQRHGIGARALKKQKVTLKKAR 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
++ +GRG +GF +I FT AK VL+ ++ K LG + V T H+LL L+ S+
Sbjct: 61 REIELYIGRG---TGFVASEIPFTPRAKRVLEMAVHEGKDLGQNFVGTEHILLALISESD 117
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
G + + G ++ K+R ++ IEEN ++ P
Sbjct: 118 GVAMRTLDKLGVNIPKLRNLILMYIEENQEEILRP------------------------- 152
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
LTQ ++ + + L+ + N++K A +GKL P +GR++++ VI+++
Sbjct: 153 -----LTQAEKFLLEREKKGSSTPT-LDEYSENISKEAVDGKLDPVIGRDKEIHEVIKVL 206
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++ +GLAQ I++ P+ L G ++ALD+ L +G
Sbjct: 207 ARRRKNNPVLIGEPGVGKTAVAEGLAQLIIAEKAPDFLDGNLLMALDLGSILAGTKYRGE 266
Query: 301 SEERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATT 359
EERI+ +++E++ +IL + E+H + A A GA A ILK AL RG +CI ATT
Sbjct: 267 FEERIKRIVEEVQNDSAIILVIDEIHTLVGAGAAEGAVDAANILKPALARGKFRCIGATT 326
Query: 360 VNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAAN 419
++E+R ++E D L+R FQPV V EP+V TIEIL GLR +E H+ L Y D+A+ AA
Sbjct: 327 IDEYRKYIERDPALERRFQPVHVKEPTVGVTIEILLGLRSKFEEHHTLSYHDKAVEQAAI 386
Query: 420 LSQQYVSERFLPDKAIDLIDEAGSHVQLCHAKVRNG 455
L+ +++++RFLPDKAID++DEAGS V+L + ++ G
Sbjct: 387 LADKFIADRFLPDKAIDVLDEAGSRVRLENRRLPRG 422
>HLYB_TREHY (Q54316) Hemolysin B
Length = 828
Score = 299 bits (766), Expect = 9e-81
Identities = 172/447 (38%), Positives = 270/447 (59%), Gaps = 41/447 (9%)
Query: 4 NLTECAKKVI-MAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSREQ 62
+LT AKKVI + AQ+EA+ + H ++ EHILLG+L +S LA ++L ID + + +
Sbjct: 5 HLTSKAKKVIELYAQEEAKRLNHDMVTPEHILLGLLYESEALATRVLMRLKIDLDRLKLE 64
Query: 63 VMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGT 122
+ + + F + + ++ S + A++L ++ + T HLLLGLL+ +GT
Sbjct: 65 LESAMVKSSTTKVFG--TLPTAPRVQKLISRSAEEARALSHNYIGTEHLLLGLLREESGT 122
Query: 123 VTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQIK 182
++ + G ++ +R+++++ + G N+++ S
Sbjct: 123 AYNVLTSMGLELTILRQEILKML-------------------GVAGSNISSMEQTS---- 159
Query: 183 FPVLTQNNNLGARKDDNANKQKS-ALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIIC 241
++DN K K+ L+ F +LTK+A++ L +GRE +V RV+QI+
Sbjct: 160 -------------QEDNVKKVKTPTLDQFARDLTKMARDKALDRVIGRENEVMRVVQILS 206
Query: 242 RRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSS 301
RR KNNP L+GEPGVGKT+I++GLA++I++ VP+ L K+V+ LD++ + +G
Sbjct: 207 RRKKNNPILLGEPGVGKTAIVEGLAEKIVAADVPDILLKKRVLTLDLSSVVAGTKYRGEF 266
Query: 302 EERIRCLIKEIELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTV 360
EERI+ ++ EI+ N+I+F+ E+H + A GA A +LK AL RG IQCI ATT+
Sbjct: 267 EERIKNIVLEIKKASNIIIFIDELHTLIGAGGAEGALDAANMLKPALSRGEIQCIGATTI 326
Query: 361 NEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANL 420
NE++ ++E D L R FQP+ V EPS+E+TIEIL G++G YE H+K+ YTDEA+ AAA L
Sbjct: 327 NEYKKYIEKDGALVRRFQPINVEEPSIEDTIEILNGIKGKYEEHHKVKYTDEAINAAAVL 386
Query: 421 SQQYVSERFLPDKAIDLIDEAGSHVQL 447
S++Y+ ER LPDKAIDLIDEAGS +L
Sbjct: 387 SKRYIFERHLPDKAIDLIDEAGSRARL 413
>CLPB_RHOBA (Q7UM33) Chaperone clpB
Length = 881
Score = 277 bits (708), Expect = 5e-74
Identities = 160/398 (40%), Positives = 236/398 (59%), Gaps = 32/398 (8%)
Query: 82 RFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQV 141
+ T A+NV+ + A S G +D +H+L + +G T L++ DV K++ +
Sbjct: 7 KLTTQAQNVVAEAQAQATSAGNAEIDPLHVLSAAVNQRDGITTPLLEKINVDVPKLKSLL 66
Query: 142 IRQIEENVHQVNLPAEGSKNQMGGQVEENVT-AESSKS--------------------NQ 180
++E+ H + ++ +E + T AES K N
Sbjct: 67 TSELEKLPHASGMGQARVSAKLQAALEASATSAESLKDEYVSTEHLLVGLARTDNKAKNL 126
Query: 181 IKFPVLTQNNNLGARK---------DDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREE 231
+ ++ N+ L A D NA ALE FG +LT+LAQ GKL P +GR+
Sbjct: 127 LSLLGVSDNDLLTAMSQIRGSARVTDPNAESTYQALEKFGIDLTQLAQSGKLDPVIGRDN 186
Query: 232 QVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADF 291
++ RVIQ++ RR KNNP L+G+PGVGKT+I +GLA RI G VP+ LKGKKVV+LD+
Sbjct: 187 EIRRVIQVLSRRTKNNPVLIGQPGVGKTAIAEGLALRIFEGDVPQSLKGKKVVSLDMGAL 246
Query: 292 LYVISNQGSSEERIRCLIKEI-ELCGNVILFVKEVHHIFDAATS-GARSFAYILKHALER 349
+ +G EER++ +++E+ + G VILF+ E+H + A + G+ A +LK L R
Sbjct: 247 VAGAKFRGDFEERLKSVLREVKDSDGKVILFIDELHLVVGAGNAEGSADAANLLKPELAR 306
Query: 350 GVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHY 409
G ++CI ATT++E+R H+E D L+R FQPV V EP+VE+T+ IL+GL+ YESH+ +
Sbjct: 307 GALRCIGATTLDEYRQHIEKDAALERRFQPVFVGEPNVEDTVAILRGLKPRYESHHGVRI 366
Query: 410 TDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
TD ALVAAANLS +Y+++RFLPDKAIDLIDEA S + +
Sbjct: 367 TDSALVAAANLSDRYIADRFLPDKAIDLIDEAASRLAM 404
>ERD1_ARATH (P42762) ERD1 protein, chloroplast precursor
Length = 945
Score = 263 bits (671), Expect = 9e-70
Identities = 160/463 (34%), Positives = 250/463 (53%), Gaps = 40/463 (8%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
+ E TE A + I+ +QKEA+ +G + T+H+LLG++ + + GI + +R
Sbjct: 79 VFERFTERAIRAIIFSQKEAKSLGKDMVYTQHLLLGLIAEDRDPQGFL--GSGITIDKAR 136
Query: 61 EQVMKLVGRGGG---------CSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHL 111
E V + S D+ F+ K V + ++++++++ + H+
Sbjct: 137 EAVWSIWDEANSDSKQEEASSTSYSKSTDMPFSISTKRVFEAAVEYSRTMDCQYIAPEHI 196
Query: 112 LLGLLQGSNGTVTQLIQNQGADVNKIREQVIRQIEENVHQVNL-PAEGSKNQMGGQVEEN 170
+GL +G+ ++++ GA++N + + +++ + + P+ SK
Sbjct: 197 AVGLFTVDDGSAGRVLKRLGANMNLLTAAALTRLKGEIAKDGREPSSSSKGSF------- 249
Query: 171 VTAESSKSNQIKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGRE 230
ES S +I A K K+ LE F +LT A EG + P +GRE
Sbjct: 250 ---ESPPSGRI------------AGSGPGGKKAKNVLEQFCVDLTARASEGLIDPVIGRE 294
Query: 231 EQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVAD 290
++V+RVIQI+CRR KNNP L+GE GVGKT+I +GLA I S P L K++++LD+
Sbjct: 295 KEVQRVIQILCRRTKNNPILLGEAGVGKTAIAEGLAISIAEASAPGFLLTKRIMSLDIGL 354
Query: 291 FLYVISNQGSSEERIRCLIKEIELCGNVILFVKEVHHIFDAATSG------ARSFAYILK 344
+ +G E R+ LI E++ G VILF+ EVH + + T G A +LK
Sbjct: 355 LMAGAKERGELEARVTALISEVKKSGKVILFIDEVHTLIGSGTVGRGNKGSGLDIANLLK 414
Query: 345 HALERGVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESH 404
+L RG +QCI +TT++E R E D L R FQPV + EPS E+ ++IL GLR YE+H
Sbjct: 415 PSLGRGELQCIASTTLDEFRSQFEKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAH 474
Query: 405 YKLHYTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
+ YT EA+ AA LS +Y+++RFLPDKAIDLIDEAGS ++
Sbjct: 475 HNCKYTMEAIDAAVYLSSRYIADRFLPDKAIDLIDEAGSRARI 517
>CLPB_PSEPK (Q88Q71) Chaperone clpB
Length = 854
Score = 260 bits (664), Expect = 6e-69
Identities = 154/394 (39%), Positives = 241/394 (61%), Gaps = 37/394 (9%)
Query: 91 LDFSLKHAKSL--GYDN--VDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQVIRQIE 146
L ++ A+SL G D+ ++ +HLL LL+ G++ L+ G D+N +R+ ++++++
Sbjct: 10 LQLAISDAQSLAVGMDHPAIEPVHLLQALLEQQGGSIKPLLMQVGFDINGLRQGLVKELD 69
Query: 147 E------NVHQVNLP---------AEGSKNQMGGQV---EENVTAESSKSNQIKFPVLTQ 188
+ VN+ A+ Q G Q E + A +++++ +L+Q
Sbjct: 70 QLPKIQNPTGDVNMSQDLARLLNQADRLAQQKGDQFISSELVLLAAMDENSKLGKLLLSQ 129
Query: 189 N----------NNL---GARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVER 235
NNL A D NA + + AL+ + +LTK A+EGKL P +GR++++ R
Sbjct: 130 GVSKKALENAINNLRGGAAVNDANAEESRQALDKYTVDLTKRAEEGKLDPVIGRDDEIRR 189
Query: 236 VIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVI 295
+Q++ RR KNNP L+GEPGVGKT+I +GLAQRI++G VP+ LKGK+++ALD+ +
Sbjct: 190 TVQVLQRRTKNNPVLIGEPGVGKTAIAEGLAQRIINGEVPDGLKGKRLLALDMGALIAGA 249
Query: 296 SNQGSSEERIRCLIKEI-ELCGNVILFVKEVHHIFDAAT-SGARSFAYILKHALERGVIQ 353
+G EER++ L+ E+ + G +ILF+ E+H + A GA +LK AL RG +
Sbjct: 250 KYRGEFEERLKSLLNELSKQEGQIILFIDELHTMVGAGKGEGAMDAGNMLKPALARGELH 309
Query: 354 CIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEA 413
C+ ATT+NE+R +E D L+R FQ V V EPS E+TI IL+GL+ YE H+K+ TD A
Sbjct: 310 CVGATTLNEYRQFIEKDAALERRFQKVLVEEPSEEDTIAILRGLKERYEVHHKVAITDGA 369
Query: 414 LVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
++AAA LS +Y+++R LPDKAIDLIDEA S +++
Sbjct: 370 IIAAAKLSHRYITDRQLPDKAIDLIDEAASRIRM 403
>CLPB_RICCN (Q92JK8) Chaperone clpB
Length = 857
Score = 259 bits (663), Expect = 8e-69
Identities = 158/400 (39%), Positives = 235/400 (58%), Gaps = 34/400 (8%)
Query: 82 RFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQV 141
+FT AK+V+ S A + + +HLL LL G + LI N G ++N +++QV
Sbjct: 5 KFTAHAKSVIASSQLLAAKNDHQQILPLHLLSSLLSEETGIIQTLINNTGGNINLLKDQV 64
Query: 142 IRQIE-------ENVHQVNLPAEGSK--NQMGGQVEEN----VTAES-----SKSNQIKF 183
++ E QV AE K + ++N VT E + N I
Sbjct: 65 QLELNKIPKVQVEGGGQVYSSAEALKVLEKASSIAKDNGDSFVTIERIFEALTYDNTIAG 124
Query: 184 PVLTQN--NNL-----------GARKD-DNANKQKSALEIFGTNLTKLAQEGKLHPFVGR 229
+LT N NN G + D ++A AL+ +G ++T+LA+ GKL P +GR
Sbjct: 125 KILTNNGINNKKIATAILQFRKGKKADTESAENSYDALKKYGRDVTELAESGKLDPIIGR 184
Query: 230 EEQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVA 289
+E++ R +Q++ RRMKNNP L+GEPGVGKT+II+GLAQRI S VPE L +++ LD+
Sbjct: 185 DEEIRRTVQVLSRRMKNNPVLIGEPGVGKTAIIEGLAQRIFSKDVPESLINCRIIELDMG 244
Query: 290 DFLYVISNQGSSEERIRCLIKEI-ELCGNVILFVKEVHHIFDAA-TSGARSFAYILKHAL 347
+ +G EER++ ++ EI E G +ILF+ E+H + T GA + +LK L
Sbjct: 245 ALIAGAKYRGEFEERLKAVLSEIKESSGEIILFIDELHLLVGTGKTDGAMDASNLLKPML 304
Query: 348 ERGVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKL 407
RG + CI ATT++E+R ++E D L R FQPV V EP+VE+TI IL+G++ YE H+ +
Sbjct: 305 ARGELHCIGATTLDEYRKYIEKDAALARRFQPVYVSEPTVEDTISILRGIKEKYELHHAV 364
Query: 408 HYTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
+D A+VAAA LS +Y+++R+LPDKAIDLIDEA S +++
Sbjct: 365 RISDSAIVAAATLSNRYITDRYLPDKAIDLIDEACSRMKI 404
>CLPB_PSESM (Q889C2) Chaperone clpB
Length = 854
Score = 256 bits (653), Expect = 1e-67
Identities = 152/399 (38%), Positives = 237/399 (59%), Gaps = 33/399 (8%)
Query: 82 RFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIREQV 141
R T + L S A L + ++ HL+ LL+ G++ L+ G D+N +R+++
Sbjct: 5 RLTSKLQLALSDSQSLAVGLDHPAIEPAHLMQALLEQQGGSIKPLLLQVGFDINSLRKEL 64
Query: 142 ------IRQIEENVHQVNLP---------AEGSKNQMGGQV---EENVTAESSKSNQIKF 183
+ +I+ VN+ A+ Q G Q E + A +++++
Sbjct: 65 SAELDRLPKIQNPTGDVNMSQDLARLLNQADRLAQQKGDQFISSELVLLAAMDENSKLGK 124
Query: 184 PVLTQN----------NNL---GARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGRE 230
+L Q NNL GA D N + + AL+ + +LTK A+EGKL P +GR+
Sbjct: 125 LLLGQGVSKKALENAINNLRGEGAVNDPNIEESRQALDKYTVDLTKRAEEGKLDPVIGRD 184
Query: 231 EQVERVIQIICRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVAD 290
+++ R IQ++ RR KNNP L+GEPGVGKT+I +GLAQRI++G VP+ L+GK++++LD+
Sbjct: 185 DEIRRTIQVLQRRTKNNPVLIGEPGVGKTAIAEGLAQRIINGEVPDGLRGKRLLSLDMGA 244
Query: 291 FLYVISNQGSSEERIRCLIKEI-ELCGNVILFVKEVHHIFDAAT-SGARSFAYILKHALE 348
+ +G EER++ L+ E+ + G +ILF+ E+H + A G+ +LK AL
Sbjct: 245 LIAGAKYRGEFEERLKSLLNELSKQEGQIILFIDELHTMVGAGKGEGSMDAGNMLKPALA 304
Query: 349 RGVIQCIFATTVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLH 408
RG + C+ ATT+NE+R ++E D L+R FQ V V EPS E+TI IL+GL+ YE H+K+
Sbjct: 305 RGELHCVGATTLNEYRQYIEKDAALERRFQKVLVDEPSEEDTIAILRGLKERYEVHHKVA 364
Query: 409 YTDEALVAAANLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
TD A++AAA LS +Y+++R LPDKAIDLIDEA S +++
Sbjct: 365 ITDGAIIAAAKLSHRYITDRQLPDKAIDLIDEAASRIRM 403
>CLB1_SYNEL (Q8DJ40) Chaperone clpB 1
Length = 871
Score = 256 bits (653), Expect = 1e-67
Identities = 150/382 (39%), Positives = 233/382 (60%), Gaps = 33/382 (8%)
Query: 98 AKSLGYDNVDTMHLLLGLLQGSNGTVTQLIQNQGADVNKIR----EQVIRQ--------- 144
AK + N+++ HL+ LL+ G TQ+ Q G V +IR E + RQ
Sbjct: 24 AKQAQHQNLESEHLMKSLLE-QEGLATQIFQKAGCSVQRIRDLTDEFISRQPKISHPSGV 82
Query: 145 -IEENVHQVNLPAEGSKNQMGGQ---VEENVTA-------------ESSKSNQIKFPVLT 187
+ +++ ++ AE ++ Q G + +E V A + S ++ +
Sbjct: 83 YLGQSLDKLLDRAEEARKQFGDEFISIEHLVLAFAQDDRFGKKLFQDIGLSEKVLREAIQ 142
Query: 188 QNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQIICRRMKNN 247
Q D N + +ALE +G +LT LA++GKL P +GR++++ RVIQI+ RR KNN
Sbjct: 143 QIRGSQKVTDQNPEGKYAALEKYGRDLTLLARQGKLDPVIGRDDEIRRVIQILSRRTKNN 202
Query: 248 PCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGSSEERIRC 307
P L+GEPGVGKT+I +GLAQRI++ VP+ L+ ++++ALD+ + +G EER++
Sbjct: 203 PVLIGEPGVGKTAIAEGLAQRIVARDVPDSLRDRQLIALDMGALIAGAKYRGEFEERLKA 262
Query: 308 LIKEI-ELCGNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFATTVNEHRM 365
++KE+ + G +ILF+ E+H + A AT GA +LK L RG ++CI ATT++E+R
Sbjct: 263 VLKEVTDSNGQIILFIDEIHTVVGAGATQGAMDAGNLLKPMLARGELRCIGATTLDEYRK 322
Query: 366 HMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAANLSQQYV 425
++E D L+R FQ V V +PSVE+TI IL+GL+ YE H+ + +D ALVAAA LS +Y+
Sbjct: 323 YIEKDAALERRFQQVYVDQPSVEDTISILRGLKERYEIHHGVKISDTALVAAATLSARYI 382
Query: 426 SERFLPDKAIDLIDEAGSHVQL 447
S+RFLPDKAIDL+DEA + +++
Sbjct: 383 SDRFLPDKAIDLVDEAAAKLKM 404
>CLPB_BIFLO (Q8G4X4) Chaperone clpB
Length = 889
Score = 255 bits (652), Expect = 1e-67
Identities = 156/449 (34%), Positives = 243/449 (53%), Gaps = 47/449 (10%)
Query: 1 MLENLTECAKKVIMAAQKEARFVGHKYISTEHILLGILGDSNGLAAKILKSYGIDFNVSR 60
M + T A++ + A + A G+ + T H++ +L NG+A ++++ G D
Sbjct: 1 MEQKFTTMAQEAVGDAIQSASAAGNAQVETLHVMDALLRQENGVARSLIEAAGGDPQAIG 60
Query: 61 EQVMKLVGRGGGCSGFSCKDIRFTFDAKNVLDFSLKHAKSLGYDNVDTMHLLLGLLQGSN 120
V + SG S + + + + K + +G + V T HLL+G+
Sbjct: 61 AAVRNALVALPSASGSSTSQPQASRQLTAAIAQAEKEMQQMGDEYVSTEHLLIGIAASKP 120
Query: 121 GTVTQLIQNQGADVNKIREQVIRQIEENVHQVNLPAEGSKNQMGGQVEENVTAESSKSNQ 180
++++ G +R+ V GG + AE S
Sbjct: 121 NQSAEILEKNGVTAASLRKAV------------------PGVRGGAKVTSPDAEGSYK-- 160
Query: 181 IKFPVLTQNNNLGARKDDNANKQKSALEIFGTNLTKLAQEGKLHPFVGREEQVERVIQII 240
ALE + T+LT A+EGKL P +GR++++ RVIQI+
Sbjct: 161 -------------------------ALEKYSTDLTAAAKEGKLDPVIGRDQEIRRVIQIL 195
Query: 241 CRRMKNNPCLVGEPGVGKTSIIQGLAQRILSGSVPEKLKGKKVVALDVADFLYVISNQGS 300
RR KNNP L+GEPGVGKT++++GLAQRI++G VP L+GKK+++LD+ + +G
Sbjct: 196 SRRTKNNPVLIGEPGVGKTAVVEGLAQRIVAGDVPTTLQGKKLISLDLGSMVAGSKYRGE 255
Query: 301 SEERIRCLIKEIELC-GNVILFVKEVHHIFDA-ATSGARSFAYILKHALERGVIQCIFAT 358
EER++ ++ EI+ G +I F+ E+H I A A G+ +LK L RG ++ I AT
Sbjct: 256 FEERLKSVLNEIKNADGQIITFIDEIHTIVGAGAAEGSMDAGNMLKPMLARGELRLIGAT 315
Query: 359 TVNEHRMHMENDTTLKRIFQPVKVVEPSVEETIEILKGLRGTYESHYKLHYTDEALVAAA 418
T++E+R ++E D L+R FQ V V EPSVE+TI IL+GL+ YE+H+K+ D+ALVAAA
Sbjct: 316 TLDEYRENIEKDPALERRFQQVFVGEPSVEDTIAILRGLKQRYEAHHKVTIGDDALVAAA 375
Query: 419 NLSQQYVSERFLPDKAIDLIDEAGSHVQL 447
LS +Y+S R LPDKAIDL+DEA +H+++
Sbjct: 376 TLSNRYISGRQLPDKAIDLVDEAAAHLRM 404
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,745,890
Number of Sequences: 164201
Number of extensions: 2133152
Number of successful extensions: 8776
Number of sequences better than 10.0: 255
Number of HSP's better than 10.0 without gapping: 183
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 7991
Number of HSP's gapped (non-prelim): 493
length of query: 455
length of database: 59,974,054
effective HSP length: 114
effective length of query: 341
effective length of database: 41,255,140
effective search space: 14068002740
effective search space used: 14068002740
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146751.5


