

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146747.9 - phase: 2 /pseudo
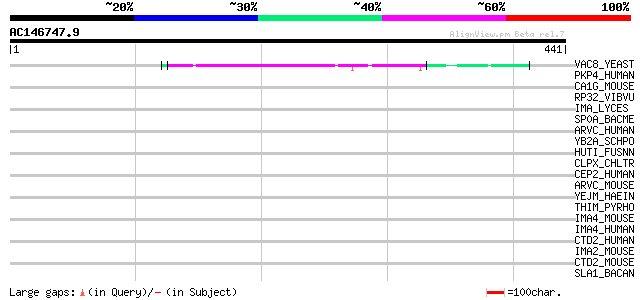
(441 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VAC8_YEAST (P39968) Vacuolar protein 8 53 1e-06
PKP4_HUMAN (Q99569) Plakophilin 4 (p0071) 37 0.078
CA1G_MOUSE (Q07563) Collagen alpha 1(XVII) chain (Bullous pemphi... 35 0.29
RP32_VIBVU (Q8DD54) RNA polymerase sigma-32 factor 34 0.66
IMA_LYCES (O22478) Importin alpha subunit (Karyopherin alpha sub... 34 0.66
SP0A_BACME (P52932) Stage 0 sporulation protein A (Fragment) 33 1.5
ARVC_HUMAN (O00192) Armadillo repeat protein deleted in velo-car... 33 1.5
YB2A_SCHPO (P87311) Hypothetical protein C31F10.10c in chromosom... 33 1.9
HUTI_FUSNN (Q8RFG1) Imidazolonepropionase (EC 3.5.2.7) (Imidazol... 33 1.9
CLPX_CHLTR (O84711) ATP-dependent Clp protease ATP-binding subun... 33 1.9
CEP2_HUMAN (Q9BV73) Centrosomal protein 2 (Centrosomal Nek2-asso... 33 1.9
ARVC_MOUSE (P98203) Armadillo repeat protein deleted in velo-car... 33 1.9
YEJM_HAEIN (P44898) Hypothetical protein HI0842 32 2.5
THIM_PYRHO (O58877) Hydroxyethylthiazole kinase (EC 2.7.1.50) (4... 32 2.5
IMA4_MOUSE (O35343) Importin alpha-4 subunit (Karyopherin alpha-... 32 2.5
IMA4_HUMAN (O00629) Importin alpha-4 subunit (Karyopherin alpha-... 32 2.5
CTD2_HUMAN (Q9UQB3) Catenin delta-2 (Delta-catenin) (Neural plak... 32 3.3
IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-... 32 4.3
CTD2_MOUSE (O35927) Catenin delta-2 (Neural plakophilin-related ... 32 4.3
SLA1_BACAN (P49051) S-layer protein sap precursor (Surface layer... 31 5.6
>VAC8_YEAST (P39968) Vacuolar protein 8
Length = 578
Score = 53.1 bits (126), Expect = 1e-06
Identities = 68/302 (22%), Positives = 121/302 (39%), Gaps = 26/302 (8%)
Query: 121 ESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESG 180
++ A+ LL L +DK+ + G + L L+ S ++ + AA++ E
Sbjct: 22 DNEREAVTLLLGYL--EDKDQLDFYSGGPLKALTTLV-YSDNLNLQRSAALAFAEITEKY 78
Query: 181 KNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGIC 240
+ E +L ++ +L S AC AL L+++ +N I GG+ L+
Sbjct: 79 VRQVSRE---VLEPILILLQSQDPQIQVAACAALGNLAVNNENKLLIVEMGGLEPLINQM 135
Query: 241 QGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANLI 300
G Q A + NLA ++ K A+I L LA S + NA G + N+
Sbjct: 136 MGDNVEVQCNAVGCITNLATRDDNKHKIATSGALIPLTKLAKSKHIRVQRNATGALLNMT 195
Query: 301 SEDESMRVLAVKEGGVECLKNYWDS---------VTMIQSLEVGVEMLRYLAMTGPIDEV 351
+E+ + L V G V L + S T + ++ V + LA T P
Sbjct: 196 HSEENRKEL-VNAGAVPVLVSLLSSTDPDVQYYCTTALSNIAVDEANRKKLAQTEP---- 250
Query: 352 LVGEGFVGRVIGVLDCDVLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGK 411
V +++ ++D V+ A A+ L + E+ G +P L+K++
Sbjct: 251 ----RLVSKLVSLMDSPSSRVKCQATLAL--RNLASDTSYQLEIVRAGGLPHLVKLIQSD 304
Query: 412 GV 413
+
Sbjct: 305 SI 306
Score = 49.3 bits (116), Expect = 2e-05
Identities = 53/212 (25%), Positives = 98/212 (46%), Gaps = 10/212 (4%)
Query: 126 AIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS-DMKEKTVAAISRVSTVESGKNNL 184
A +LL++ + ++ + A G VPVLV LL S+ D++ A+S ++ E+ + L
Sbjct: 187 ATGALLNMTHSEENRKELVNA-GAVPVLVSLLSSTDPDVQYYCTTALSNIAVDEANRKKL 245
Query: 185 LAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGICQGGT 244
L++ LV ++DS S +A +AL+ L+ I GG+ L+ + Q +
Sbjct: 246 AQTEPRLVSKLVSLMDSPSSRVKCQATLALRNLASDTSYQLEIVRAGGLPHLVKLIQSDS 305
Query: 245 PGSQGYAAAVLRNLAKFNEIKENFVEE----NAVIVLLGLASSGTGLARENAIGCVANLI 300
+ A +RN++ + + E + + ++ LL S + +A+ + NL
Sbjct: 306 IPLVLASVACIRNIS-IHPLNEGLIVDAGFLKPLVRLLDYKDSEE--IQCHAVSTLRNLA 362
Query: 301 SEDESMRVLAVKEGGVE-CLKNYWDSVTMIQS 331
+ E R + G VE C + DS +QS
Sbjct: 363 ASSEKNRKEFFESGAVEKCKELALDSPVSVQS 394
>PKP4_HUMAN (Q99569) Plakophilin 4 (p0071)
Length = 1211
Score = 37.4 bits (85), Expect = 0.078
Identities = 34/133 (25%), Positives = 60/133 (44%), Gaps = 13/133 (9%)
Query: 150 VPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAI-- 207
+P+LV LL +D + A R ++ L+ G + LV L G+G ++
Sbjct: 874 LPILVELLRMDNDRVVSSGATALRNMALDVRNKELI--GKYAMRDLVNRLPGGNGPSVLS 931
Query: 208 ----EKACIAL-QALSLSRDNARAIGSRGGISSLLGICQGGTPGSQ----GYAAAVLRNL 258
C AL + S + +NA+A+ GGI L+ I +G S AA VL L
Sbjct: 932 DETMAAICCALHEVTSKNMENAKALADSGGIEKLVNITKGRGDRSSLKVVKAAAQVLNTL 991
Query: 259 AKFNEIKENFVEE 271
++ +++ + ++
Sbjct: 992 WQYRDLRSIYKKD 1004
>CA1G_MOUSE (Q07563) Collagen alpha 1(XVII) chain (Bullous
pemphigoid antigen 2) (180 kDa bullous pemphigoid
antigen 2)
Length = 1470
Score = 35.4 bits (80), Expect = 0.29
Identities = 37/154 (24%), Positives = 67/154 (43%), Gaps = 13/154 (8%)
Query: 95 TPAFSKREAVRSLTRN--------LIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVA 146
+P + ++E S TR + RLQ SP +R T +D + SLL + ++ + +
Sbjct: 119 SPEYPRKELASSSTRGRSQTRESEIRVRLQSASPSTRWTELDEVKSLL-KGSRSASASPT 177
Query: 147 QGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLA 206
+ L + ++ KTV A S +V ++ + + + L +GS L
Sbjct: 178 RNTSNTLP--IPKKGTVETKTVTASSH--SVSGTYDSAILDTNFPPHMWSSTLPAGSSLG 233
Query: 207 IEKACIALQALSLSRDNARAIGSRGGISSLLGIC 240
+ I Q+ SL NA + GS G+ + + C
Sbjct: 234 TYQNNITAQSTSLLNTNAYSTGSVFGVPNNMASC 267
>RP32_VIBVU (Q8DD54) RNA polymerase sigma-32 factor
Length = 285
Score = 34.3 bits (77), Expect = 0.66
Identities = 32/99 (32%), Positives = 49/99 (49%), Gaps = 15/99 (15%)
Query: 104 VRSLTRNL-IARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSD 162
V ++ R L + ++ ESR A+DS L N++D + +++ A PVL L D SSD
Sbjct: 154 VETVARELGVEPAEVREMESRLAAVDSTFDLPNEEDDSSSVSTA----PVLY-LEDKSSD 208
Query: 163 MKEKTVAAISRVSTVESGKNNLLAEGLLLLN----HLVR 197
+ + A E+ NN LA L L+ H+VR
Sbjct: 209 VADNLEA-----QNWEAHTNNRLAMALASLDERSQHIVR 242
>IMA_LYCES (O22478) Importin alpha subunit (Karyopherin alpha
subunit) (KAP alpha)
Length = 527
Score = 34.3 bits (77), Expect = 0.66
Identities = 67/357 (18%), Positives = 150/357 (41%), Gaps = 67/357 (18%)
Query: 120 PESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDS-SSDMKEKTVAAISRVS--- 175
P+ + A +L ++ + +N + + G VP+ +RLL S S D++E+ V A+ ++
Sbjct: 131 PQLQFEAAWALTNIASGTSENTKVVIDYGSVPIFIRLLSSPSDDVREQAVWALGNIAGDS 190
Query: 176 -------------------TVESGKNNLLAEGLLLLNH-------------------LVR 197
E K ++L L++ L R
Sbjct: 191 PKYRDLVLGHGALVALLAQFNEQAKLSMLRNATWTLSNFCRGKPQPLFEQTKAALPTLGR 250
Query: 198 VLDSGSGLAIEKACIALQALS-LSRDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLR 256
++ S + AC AL LS + D +A+ G S L+ + +P A +
Sbjct: 251 LIHSNDEEVLTDACWALSYLSDGTNDKIQAVIEAGVCSRLVELLLHSSPSVLIPALRTVG 310
Query: 257 NLAKFNEIK-ENFVEENAVIVLLGLASSGTGLA-RENAIGCVANLISEDESMRVLAVKEG 314
N+ ++I+ + ++ +A+ L+ L + + ++ A ++N+ + + + + ++ G
Sbjct: 311 NIVTGDDIQTQVMIDHHALPCLVNLLTQNYKKSIKKEACWTISNITAGNRNQIQIVIEAG 370
Query: 315 GVECLKNYWDSVTMIQSLEVGVEMLRYLAMT-----GPIDEV--LVGEGFVGRVIGVLDC 367
+ L V ++Q+ E ++ A++ G D++ LV +G + + +L C
Sbjct: 371 IIAPL------VYLLQNAEFEIKKEAAWAISNATSGGNHDQIKFLVSQGCIKPLCDLLVC 424
Query: 368 D---VLTVRIAAARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLD-GKGVEEKDLLQ 420
++TV + + +G + K++G V +++D +G+E+ + LQ
Sbjct: 425 PDPRIVTVCLEGLENILKIG-----EADKDLGNTEGVNVYAQLIDEAEGLEKIENLQ 476
>SP0A_BACME (P52932) Stage 0 sporulation protein A (Fragment)
Length = 212
Score = 33.1 bits (74), Expect = 1.5
Identities = 20/59 (33%), Positives = 30/59 (49%)
Query: 293 IGCVANLISEDESMRVLAVKEGGVECLKNYWDSVTMIQSLEVGVEMLRYLAMTGPIDEV 351
IG + + ISE E M V+ V G ECL D + L++ + L LA+ G + E+
Sbjct: 16 IGLLEDYISEQEDMEVIGVAYNGQECLSILKDKNPDVLVLDIIMPHLDGLAVLGQLKEM 74
>ARVC_HUMAN (O00192) Armadillo repeat protein deleted in
velo-cardio-facial syndrome
Length = 962
Score = 33.1 bits (74), Expect = 1.5
Identities = 37/146 (25%), Positives = 59/146 (40%), Gaps = 21/146 (14%)
Query: 195 LVRVLDSGSGLAIEKACIALQALSLSR--DNARAIGSRGGISSLLGICQGGTPGS-QGYA 251
LV +LD +AC AL+ LS R DN AI GG+ +L+ + + +
Sbjct: 405 LVALLDHPRAEVRRRACGALRNLSYGRDTDNKAAIRDCGGVPALVRLLRAARDNEVRELV 464
Query: 252 AAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARE-----------------NAIG 294
L NL+ + +K ++ + + +G RE N G
Sbjct: 465 TGTLWNLSSYEPLKMVIIDHGLQTLTHEVIVPHSGWEREPNEDSKPRDAEWTTVFKNTSG 524
Query: 295 CVANLISED-ESMRVLAVKEGGVECL 319
C+ N+ S+ E+ R L EG V+ L
Sbjct: 525 CLRNVSSDGAEARRRLRECEGLVDAL 550
>YB2A_SCHPO (P87311) Hypothetical protein C31F10.10c in chromosome
II
Length = 574
Score = 32.7 bits (73), Expect = 1.9
Identities = 25/86 (29%), Positives = 41/86 (47%), Gaps = 7/86 (8%)
Query: 331 SLEVGVEMLRYLAMTGP-IDEVLVGEGFVGRVIGVLDCDV------LTVRIAAARAVYAM 383
SL + L YL T P I E+L +G + R++ +L +T+ A + V +
Sbjct: 35 SLMNSLSHLVYLTSTSPKIREILTMDGGLMRLMNILRAGRGQTFARMTIWQLALQCVVNV 94
Query: 384 GLNGGNKTRKEMGECGCVPSLIKMLD 409
G+ G R + E G VP ++ +LD
Sbjct: 95 GIRGSEAIRIRVVEAGIVPIVVTLLD 120
>HUTI_FUSNN (Q8RFG1) Imidazolonepropionase (EC 3.5.2.7)
(Imidazolone-5-propionate hydrolase)
Length = 413
Score = 32.7 bits (73), Expect = 1.9
Identities = 56/220 (25%), Positives = 92/220 (41%), Gaps = 39/220 (17%)
Query: 127 IDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRV-STVESGKNNLL 185
IDS L++ + A+ VP L ++ EK +S + ST + + L+
Sbjct: 76 IDSHTHLVHGGSRENEFAMKIAGVPYL--------EILEKGGGILSTLKSTRNASEQELI 127
Query: 186 AEGLLLLNHLVRV------LDSGSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLGI 239
+ L L H++ + SG GL +E L+ ++ +LG
Sbjct: 128 EKTLKSLRHMLELGVTTVEAKSGYGLNLEDELKQLE-----------------VTKILGY 170
Query: 240 CQGGTPGSQGYAAAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANL 299
Q T S AA ++ + KE +V+E VI +L + LA I C +
Sbjct: 171 LQPVTLVSTFMAAHATP--PEYKDNKEGYVQE--VIKMLPIVKE-RNLAEFCDIFCEDKV 225
Query: 300 ISEDESMRVLAV-KEGGVECLKNYWDSVTMIQSLEVGVEM 338
S DES R+L V KE G + LK + D + + +E+ E+
Sbjct: 226 FSVDESRRILTVAKELGYK-LKIHADEIVSLGGVELAAEL 264
>CLPX_CHLTR (O84711) ATP-dependent Clp protease ATP-binding subunit
clpX
Length = 419
Score = 32.7 bits (73), Expect = 1.9
Identities = 33/143 (23%), Positives = 64/143 (44%), Gaps = 23/143 (16%)
Query: 64 QTQSQIDSLIATLHRHVTDCDVLFRSGLLLETPAFSKREAVRSLTRNLIARLQIGSPESR 123
+++ ++ LIA ++ D + SG+L +TPA + +E S T + + L++ +P+
Sbjct: 14 RSEKDVEKLIAGPSVYICDYCIKLCSGILDKTPAPATQEIATSSTSSPTS-LRVLTPKEI 72
Query: 124 ATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNN 183
IDS + + K +++AV K + A+ + V GK+N
Sbjct: 73 KRHIDSYVIGQERAKKTISVAVYN----------------HYKRIRALMQDKQVSYGKSN 116
Query: 184 LLAEG------LLLLNHLVRVLD 200
+L G L+ L ++LD
Sbjct: 117 VLLLGPTGSGKTLIAKTLAKILD 139
>CEP2_HUMAN (Q9BV73) Centrosomal protein 2 (Centrosomal
Nek2-associated protein 1) (C-NAP1) (Centrosome protein
250) (Centrosome associated protein CEP250)
Length = 2442
Score = 32.7 bits (73), Expect = 1.9
Identities = 49/232 (21%), Positives = 93/232 (39%), Gaps = 42/232 (18%)
Query: 7 KLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPHGKLQTQ 66
++ +L+ LTD E S+ +H+ LQ L +S + +S+ GK Q
Sbjct: 1265 QVQKLEERLTDTEAEKSQ-----------VHTELQDLQRQLSQNQEEKSKW--EGK---Q 1308
Query: 67 SQIDSLIATLHRHVTDCDVLFRSGLLLETPAFSKREAVRSLTRNLIAR---LQIGSPESR 123
+ ++S + LH + R L A +RE +++ NL A+ LQ E+R
Sbjct: 1309 NSLESELMELHETMASLQSRLRRAELQRMEAQGERELLQAAKENLTAQVEHLQAAVVEAR 1368
Query: 124 ATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNN 183
A A S +L +D + A ++L + + + + A+ ++ +
Sbjct: 1369 AQA--SAAGILEEDLRTARSA---------LKLKNEEVESERERAQALQEQGELKVAQGK 1417
Query: 184 LLAEGLLLLNHLV------------RVLDSGSGLAIEKACIALQALSLSRDN 223
L E L LL + ++ + ++KA + L +L L + N
Sbjct: 1418 ALQENLALLTQTLAEREEEVETLRGQIQELEKQREMQKAALELLSLDLKKRN 1469
>ARVC_MOUSE (P98203) Armadillo repeat protein deleted in
velo-cardio-facial syndrome homolog (Fragment)
Length = 969
Score = 32.7 bits (73), Expect = 1.9
Identities = 37/146 (25%), Positives = 58/146 (39%), Gaps = 21/146 (14%)
Query: 195 LVRVLDSGSGLAIEKACIALQALSLSR--DNARAIGSRGGISSLLGICQGGTPGS-QGYA 251
LV +LD +AC AL+ LS R DN AI GG+ L+ + + +
Sbjct: 400 LVALLDHPRAEVRRRACGALRNLSYGRDTDNKAAIRDCGGVPXLVRLLRAARDNEVRELV 459
Query: 252 AAVLRNLAKFNEIKENFVEENAVIVLLGLASSGTGLARE-----------------NAIG 294
L NL+ + +K ++ + + +G RE N G
Sbjct: 460 TGTLWNLSSYEPLKMVIIDHGLQTLTHEVIVPHSGWEREPNEDSKPRDAEWTTVFKNTSG 519
Query: 295 CVANLISED-ESMRVLAVKEGGVECL 319
C+ N+ S+ E+ R L EG V+ L
Sbjct: 520 CLRNVSSDGAEARRRLRECEGLVDAL 545
>YEJM_HAEIN (P44898) Hypothetical protein HI0842
Length = 585
Score = 32.3 bits (72), Expect = 2.5
Identities = 40/159 (25%), Positives = 72/159 (45%), Gaps = 16/159 (10%)
Query: 7 KLTQLQTHLTDFSTEYSKSSTTNPLSLHLLHSVLQTLNDAVSLSHHCQSQILPHGKLQTQ 66
KL + T T+F+ YS ++ N + L + + D++ LS+H QS ++ KL+ +
Sbjct: 287 KLAKFATSSTEFTNHYSTGNSNNAGLIGLFYGLNANYTDSI-LSNHTQSVLIE--KLRAE 343
Query: 67 S-QIDSLIATLHRHVTDCDVLFRSGLLLETPAFSKREAV---RSLTRNLIARLQIGSPES 122
+ Q+ AT + D +FR L E S + S +NL ++ +S
Sbjct: 344 NYQLGLFSATNFK-----DSIFRQALFREIKLSSNKTNKPNNESAVKNLNDFIKAQKTDS 398
Query: 123 RATA-IDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSS 160
A +D L N D + T+ Q + +L + L+S+
Sbjct: 399 PWFAYLDLALEAKNPSDYDRTL---QDIDSLLAKALEST 434
>THIM_PYRHO (O58877) Hydroxyethylthiazole kinase (EC 2.7.1.50)
(4-methyl-5-beta-hydroxyethylthiazole kinase) (Thz
kinase) (TH kinase)
Length = 265
Score = 32.3 bits (72), Expect = 2.5
Identities = 27/97 (27%), Positives = 47/97 (47%), Gaps = 5/97 (5%)
Query: 145 VAQGVVPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVR--VLDS- 201
+A G PV+ + +M A + + T++SG + + + N L + VLD
Sbjct: 35 LALGASPVMAHAEEELEEMIRLADAVVINIGTLDSGWRRSMVKATEIANELGKPIVLDPV 94
Query: 202 GSGLAIEKACIALQALSLSRDNARAIGSRGGISSLLG 238
G+G + ++L+ LS D + G+ G IS+LLG
Sbjct: 95 GAGATKFRTRVSLEILSRGVDVLK--GNFGEISALLG 129
>IMA4_MOUSE (O35343) Importin alpha-4 subunit (Karyopherin alpha-4
subunit) (Importin alpha Q1)
Length = 521
Score = 32.3 bits (72), Expect = 2.5
Identities = 49/218 (22%), Positives = 94/218 (42%), Gaps = 11/218 (5%)
Query: 204 GLAIEKACIALQALSLSRDNARAIGSRGGISSLLGIC--QGGTPGSQGYAAAVLRNLAK- 260
G+ + A + LS R+ + GI +L C + P Q AA L N+A
Sbjct: 86 GIQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVHCLERDDNPSLQFEAAWALTNIASG 145
Query: 261 FNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLK 320
+E + V+ NAV + L L S E A+ + N+I + R + G V+ L
Sbjct: 146 TSEQTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLL 205
Query: 321 NYWD---SVTMIQSLE-VGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAA 376
++ +T ++++ V V + R+ P++ + + +I D ++L + A
Sbjct: 206 SFISPSIPITFLRNVTWVMVNLCRHKDPPPPMETIQEILPALCVLIHHTDVNILVDTVWA 265
Query: 377 ARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGKGVE 414
+ + GN+ + + + G VP L+ +L + V+
Sbjct: 266 LSYL----TDAGNEQIQMVIDSGIVPHLVPLLSHQEVK 299
>IMA4_HUMAN (O00629) Importin alpha-4 subunit (Karyopherin alpha-4
subunit) (Qip1 protein)
Length = 521
Score = 32.3 bits (72), Expect = 2.5
Identities = 49/218 (22%), Positives = 94/218 (42%), Gaps = 11/218 (5%)
Query: 204 GLAIEKACIALQALSLSRDNARAIGSRGGISSLLGIC--QGGTPGSQGYAAAVLRNLAK- 260
G+ + A + LS R+ + GI +L C + P Q AA L N+A
Sbjct: 86 GIQLSAVQAARKLLSSDRNPPIDDLIKSGILPILVHCLERDDNPSLQFEAAWALTNIASG 145
Query: 261 FNEIKENFVEENAVIVLLGLASSGTGLARENAIGCVANLISEDESMRVLAVKEGGVECLK 320
+E + V+ NAV + L L S E A+ + N+I + R + G V+ L
Sbjct: 146 TSEQTQAVVQSNAVPLFLRLLHSPHQNVCEQAVWALGNIIGDGPQCRDYVISLGVVKPLL 205
Query: 321 NYWD---SVTMIQSLE-VGVEMLRYLAMTGPIDEVLVGEGFVGRVIGVLDCDVLTVRIAA 376
++ +T ++++ V V + R+ P++ + + +I D ++L + A
Sbjct: 206 SFISPSIPITFLRNVTWVMVNLCRHKDPPPPMETIQEILPALCVLIHHTDVNILVDTVWA 265
Query: 377 ARAVYAMGLNGGNKTRKEMGECGCVPSLIKMLDGKGVE 414
+ + GN+ + + + G VP L+ +L + V+
Sbjct: 266 LSYL----TDAGNEQIQMVIDSGIVPHLVPLLSHQEVK 299
>CTD2_HUMAN (Q9UQB3) Catenin delta-2 (Delta-catenin) (Neural
plakophilin-related ARM-repeat protein) (NPRAP)
(Neurojungin) (GT24)
Length = 1225
Score = 32.0 bits (71), Expect = 3.3
Identities = 32/137 (23%), Positives = 59/137 (42%), Gaps = 18/137 (13%)
Query: 151 PVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSG--SGLAIE 208
P+LV LL +D VA R ++ L+ G + LV L G S
Sbjct: 895 PILVELLRIDNDRVACAVATALRNMALDVRNKELI--GKYAMRDLVHRLPGGNNSNNTAS 952
Query: 209 KA----------CIALQALSLSRDNARAIGSRGGISSLLGICQG----GTPGSQGYAAAV 254
KA C + ++ + +NA+A+ GGI L+GI + +P A+ V
Sbjct: 953 KAMSDDTVTAVCCTLHEVITKNMENAKALRDAGGIEKLVGISKSKGDKHSPKVVKAASQV 1012
Query: 255 LRNLAKFNEIKENFVEE 271
L ++ ++ +++ + ++
Sbjct: 1013 LNSMWQYRDLRSLYKKD 1029
>IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin)
(Pore targeting complex 58 kDa subunit) (PTAC58)
(Importin alpha P1)
Length = 529
Score = 31.6 bits (70), Expect = 4.3
Identities = 39/225 (17%), Positives = 92/225 (40%), Gaps = 9/225 (4%)
Query: 102 EAVRSLTRNLIARLQIGSPESRATAIDSLLSLLNQDDKNVTIAVAQGVVPVLVRLLDSSS 161
+AV + L+ L PE A + ++ L + ++ + + V +GVVP LV+LL ++
Sbjct: 247 DAVEQILPTLVRLLHHNDPEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVKLLGATE 306
Query: 162 -DMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGLAIEKACIALQALSLS 220
+ + AI + T + + + L + + + + E +
Sbjct: 307 LPIVTPALRAIGNIVTGTDEQTQKVIDAGALAVFPSLLTNPKTNIQKEATWTMSNITAGR 366
Query: 221 RDNARAIGSRGGISSLLGICQGGTPGSQGYAAAVLRNLAKFNEIKE--NFVEENAVIVLL 278
+D + + + G + L+G+ +Q AA + N +++ V + L+
Sbjct: 367 QDQIQQVVNHGLVPFLVGVLSKADFKTQKEAAWAITNYTSGGTVEQIVYLVHCGIIEPLM 426
Query: 279 GLASSGTGLARENAIGCVANL------ISEDESMRVLAVKEGGVE 317
L S+ + + ++N+ + E E + ++ + GG++
Sbjct: 427 NLLSAKDTKIIQVILDAISNIFQAAEKLGETEKLSIMIEECGGLD 471
>CTD2_MOUSE (O35927) Catenin delta-2 (Neural plakophilin-related
ARM-repeat protein) (NPRAP) (Neurojungin)
Length = 1247
Score = 31.6 bits (70), Expect = 4.3
Identities = 30/138 (21%), Positives = 60/138 (42%), Gaps = 18/138 (13%)
Query: 150 VPVLVRLLDSSSDMKEKTVAAISRVSTVESGKNNLLAEGLLLLNHLVRVLDSGSGL---- 205
+P+LV LL +D VA R ++ L+ G + LV L G+
Sbjct: 916 LPILVELLRIDNDRVVCAVATALRNMALDVRNKELI--GKYAMRDLVHRLPGGNNSNNSG 973
Query: 206 -------AIEKACIAL-QALSLSRDNARAIGSRGGISSLLGICQG----GTPGSQGYAAA 253
+ C L + ++ + +NA+A+ GGI L+GI + +P A+
Sbjct: 974 SKAMSDDTVTAVCCTLHEVITKNMENAKALRDAGGIEKLVGISKSKGDKHSPKVVKAASQ 1033
Query: 254 VLRNLAKFNEIKENFVEE 271
VL ++ ++ +++ + ++
Sbjct: 1034 VLNSMWQYRDLRSLYKKD 1051
>SLA1_BACAN (P49051) S-layer protein sap precursor (Surface layer
protein) (Surface array protein)
Length = 814
Score = 31.2 bits (69), Expect = 5.6
Identities = 30/126 (23%), Positives = 59/126 (46%), Gaps = 23/126 (18%)
Query: 178 ESGKNNLLAEGLLLLNHLVRVLDSG---SGLAIEKACIALQALS------------LSRD 222
E+ K ++ +G L+ H +V+D+ GLA+E +L+ ++ LS D
Sbjct: 681 ETYKVTVVLDGKLITTHSFKVVDTAPTAKGLAVEFTSTSLKEVAPNADLKAALLNILSVD 740
Query: 223 NARAIGSRGGISSLLGI-------CQGGTPGSQGYAAAVLRNLAKFNEIKENFVE-ENAV 274
A ++ +S++ + + GT G++G + ++NL + KE VE + AV
Sbjct: 741 GVPATTAKATVSNVEFVSADTNVVAENGTVGAKGATSIYVKNLTVVKDGKEQKVEFDKAV 800
Query: 275 IVLLGL 280
V + +
Sbjct: 801 QVAVSI 806
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,311,769
Number of Sequences: 164201
Number of extensions: 1877004
Number of successful extensions: 5370
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 5340
Number of HSP's gapped (non-prelim): 63
length of query: 441
length of database: 59,974,054
effective HSP length: 113
effective length of query: 328
effective length of database: 41,419,341
effective search space: 13585543848
effective search space used: 13585543848
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146747.9


