

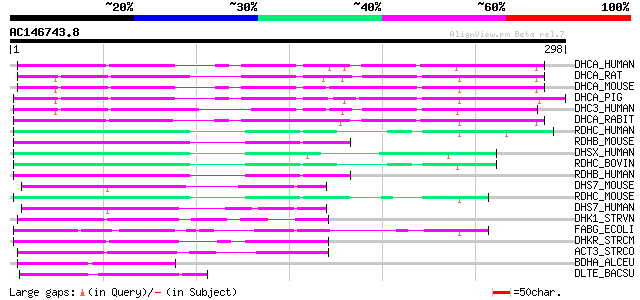
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146743.8 - phase: 0
(298 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DHCA_HUMAN (P16152) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) ... 140 4e-33
DHCA_RAT (P47727) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (N... 137 2e-32
DHCA_MOUSE (P48758) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) ... 137 4e-32
DHCA_PIG (Q28960) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (N... 135 1e-31
DHC3_HUMAN (O75828) Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) ... 134 2e-31
DHCA_RABIT (P47844) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) ... 126 6e-29
RDHC_HUMAN (Q96NR8) Retinol dehydrogenase 12 (EC 1.1.1.-) 71 3e-12
RDHB_MOUSE (Q9QYF1) Retinol dehydrogenase 11 (EC 1.1.1.-) (Retin... 65 2e-10
DHSX_HUMAN (Q8N5I4) Dehydrogenase/reductase SDR family member on... 65 2e-10
RDHC_BOVIN (P59837) Retinol dehydrogenase 12 (EC 1.1.1.-) (Doubl... 64 6e-10
RDHB_HUMAN (Q8TC12) Retinol dehydrogenase 11 (EC 1.1.1.-) (Retin... 62 2e-09
DHS7_MOUSE (Q9CXR1) Dehydrogenase/reductase SDR family member 7 ... 60 5e-09
RDHC_MOUSE (Q8BYK4) Retinol dehydrogenase 12 (EC 1.1.1.-) 60 7e-09
DHS7_HUMAN (Q9Y394) Dehydrogenase/reductase SDR family member 7 ... 59 2e-08
DHK1_STRVN (P16542) Granaticin polyketide synthase putative keto... 59 2e-08
FABG_ECOLI (P25716) 3-oxoacyl-[acyl-carrier-protein] reductase (... 57 4e-08
DHKR_STRCM (P41177) Monensin polyketide synthase putative ketoac... 57 6e-08
ACT3_STRCO (P16544) Putative ketoacyl reductase (EC 1.3.1.-) 57 6e-08
BDHA_ALCEU (Q9X6U2) D-beta-hydroxybutyrate dehydrogenase (EC 1.1... 56 1e-07
DLTE_BACSU (P39577) Protein dltE 55 2e-07
>DHCA_HUMAN (P16152) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 1)
(Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189)
(Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin
dehydrogenase [NADP+]) (EC 1.1.1.197)
Length = 276
Score = 140 bits (353), Expect = 4e-33
Identities = 106/297 (35%), Positives = 158/297 (52%), Gaps = 52/297 (17%)
Query: 5 AVVTGANKGIGLEIVKQLAFL-GVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLD 63
A+VTG NKGIGL IV+ L L VVLTAR+ TRG+ A+ +L GLS FHQLD+ D
Sbjct: 7 ALVTGGNKGIGLAIVRDLCRLFSGDVVLTARDVTRGQAAVQQLQAEGLS-PRFHQLDIDD 65
Query: 64 ALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGV 123
SI +L F++ ++G LD+L+NNAG +A KV++
Sbjct: 66 LQSIRALRDFLRKEYGGLDVLVNNAG---------------------IAFKVADP----- 99
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRG--ELKRIPNE--- 178
T + +AE + TN++G + V LLPL++ + R+VN+SS+ LK E
Sbjct: 100 -TPFHIQAEVTMKTNFFGTRDVCTELLPLIK---PQGRVVNVSSIMSVRALKSCSPELQQ 155
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLA 238
+ R+E ++E ++ ++ KF+ D K H+ GW AY ++K + +R+ A
Sbjct: 156 KFRSE-----TITEEELVGLMNKFVEDTKKGVHQKEGWPS--SAYGVTKIGVTVLSRIHA 208
Query: 239 ------KKNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
+K +L+N PG+V TD K T + +EGA PV L+LLP A+GP G
Sbjct: 209 RKLSEQRKGDKILLNACCPGWVRTDMAGPKATKSPEEGAETPVYLALLPPDAEGPHG 265
>DHCA_RAT (P47727) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 1)
Length = 276
Score = 137 bits (346), Expect = 2e-32
Identities = 103/298 (34%), Positives = 156/298 (51%), Gaps = 54/298 (18%)
Query: 5 AVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
A+VTGANKGIG IV+ L FLG VVLTAR+++RG +A+ +L GLS FHQLD+
Sbjct: 7 ALVTGANKGIGFAIVRDLCRKFLG-DVVLTARDESRGHEAVKQLQTEGLSP-RFHQLDID 64
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
+ SI +L F+ ++G L++L+NNAG + VD
Sbjct: 65 NPQSIRALRDFLLQEYGGLNVLVNNAGIAFKVVDP------------------------- 99
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSS---LRGELKRIPN-- 177
T + +AE + TN++G + V LLP+++ + R+VN+SS LR P
Sbjct: 100 --TPFHIQAEVTMKTNFFGTQDVCKELLPIIK---PQGRVVNVSSSVSLRALKSCSPELQ 154
Query: 178 ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
++ R+E ++E ++ ++ KF+ D K H GW AY ++K + +R+
Sbjct: 155 QKFRSET-----ITEEELVGLMNKFIEDAKKGVHAKEGWPNS--AYGVTKIGVTVLSRIY 207
Query: 238 AKK------NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
A+K +L+N PG+V TD K T + +EGA PV L+LLP A+GP G
Sbjct: 208 ARKLNEERREDKILLNACCPGWVRTDMAGPKATKSPEEGAETPVYLALLPPGAEGPHG 265
>DHCA_MOUSE (P48758) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 1)
Length = 276
Score = 137 bits (344), Expect = 4e-32
Identities = 104/294 (35%), Positives = 156/294 (52%), Gaps = 46/294 (15%)
Query: 5 AVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
A+VTGANKGIG I + L F G VVL AR++ RG+ A+ KL GLS FHQLD+
Sbjct: 7 ALVTGANKGIGFAITRDLCRKFSG-DVVLAARDEERGQTAVQKLQAEGLSP-RFHQLDID 64
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
+ SI +L F+ ++G LD+L+N AG +A KV++
Sbjct: 65 NPQSIRALRDFLLKEYGGLDVLVNKAG---------------------IAFKVNDD---- 99
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRN 182
T + +AE + TN++G + V LLPL++ + R+VN+SS+ L+ + N RL
Sbjct: 100 --TPFHIQAEVTMETNFFGTRDVCKELLPLIK---PQGRVVNVSSMVS-LRALKNCRLEL 153
Query: 183 ELGDVDE-LSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK- 240
+ E ++E ++ ++ KF+ D K H GW AY ++K + +R+LA+K
Sbjct: 154 QQKFRSETITEEELVGLMNKFVEDTKKGVHAEEGWPNS--AYGVTKIGVTVLSRILARKL 211
Query: 241 -----NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
+L+N PG+V TD K T + +EGA PV L+LLP A+GP G
Sbjct: 212 NEQRREDKILLNACCPGWVRTDMAGPKATKSPEEGAETPVYLALLPPDAEGPHG 265
>DHCA_PIG (Q28960) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 1)
(20-beta-hydroxysteroid dehydrogenase)
(Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189)
(Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin
dehydroge
Length = 288
Score = 135 bits (340), Expect = 1e-31
Identities = 103/309 (33%), Positives = 159/309 (51%), Gaps = 50/309 (16%)
Query: 3 RYAVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLD 60
R A+VTGANKGIG IV+ L F G VVLTAR+ RG+ A+ +L GLS FHQLD
Sbjct: 5 RVALVTGANKGIGFAIVRDLCRQFAG-DVVLTARDVARGQAAVKQLQAEGLSP-RFHQLD 62
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLL 120
++D SI +L F++ ++G LD+L+NNA +A ++ N
Sbjct: 63 IIDLQSIRALCDFLRKEYGGLDVLVNNAA---------------------IAFQLDNP-- 99
Query: 121 QGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNE-- 178
T + +AE + TN+ G + V LLPL++ + R+VN+SS G R NE
Sbjct: 100 ----TPFHIQAELTMKTNFMGTRNVCTELLPLIK---PQGRVVNVSSTEGV--RALNECS 150
Query: 179 -RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVL 237
L+ + + ++E ++ ++ KF+ D K H GW Y ++K ++ +R+
Sbjct: 151 PELQQKFKS-ETITEEELVGLMNKFVEDTKNGVHRKEGWSDS--TYGVTKIGVSVLSRIY 207
Query: 238 AKK------NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPAD--GPTGCY 289
A+K +L+N PG+V TD K + + GA PV L+LLP+D GP G +
Sbjct: 208 ARKLREQRAGDKILLNACCPGWVRTDMGGPKAPKSPEVGAETPVYLALLPSDAEGPHGQF 267
Query: 290 FDCTEIAEF 298
++ E+
Sbjct: 268 VTDKKVVEW 276
>DHC3_HUMAN (O75828) Carbonyl reductase [NADPH] 3 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 3)
Length = 276
Score = 134 bits (338), Expect = 2e-31
Identities = 103/295 (34%), Positives = 148/295 (49%), Gaps = 54/295 (18%)
Query: 3 RYAVVTGANKGIGLEIVKQLA--FLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLD 60
R A+VTGAN+GIGL I ++L F G VVLTAR+ RG+ A+ +L GLS FHQLD
Sbjct: 5 RVALVTGANRGIGLAIARELCRQFSG-DVVLTARDVARGQAAVQQLQAEGLSP-RFHQLD 62
Query: 61 VLDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLL 120
+ D SI +L F++ ++G L++L+NNA + D
Sbjct: 63 IDDLQSIRALRDFLRKEYGGLNVLVNNAAVAFKSDDPMPFDI------------------ 104
Query: 121 QGVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPN--- 177
KAE L TN++ + + LLP+++ R+VN+SSL+ L+ N
Sbjct: 105 ---------KAEMTLKTNFFATRNMCNELLPIMK---PHGRVVNISSLQC-LRAFENCSE 151
Query: 178 ---ERLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYT 234
ER +E L+EG + ++KKF+ D K HE GW Y +SK + +
Sbjct: 152 DLQERFHSET-----LTEGDLVDLMKKFVEDTKNEVHEREGWPNS--PYGVSKLGVTVLS 204
Query: 235 RVLAK------KNPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLPAD 283
R+LA+ K +L+N PG V TD + TV+EGA PV L+LLP D
Sbjct: 205 RILARRLDEKRKADRILVNACCPGPVKTDMDGKDSIRTVEEGAETPVYLALLPPD 259
>DHCA_RABIT (P47844) Carbonyl reductase [NADPH] 1 (EC 1.1.1.184)
(NADPH-dependent carbonyl reductase 1)
Length = 276
Score = 126 bits (317), Expect = 6e-29
Identities = 93/296 (31%), Positives = 150/296 (50%), Gaps = 46/296 (15%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFL-GVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDV 61
R A+VTGANKG+G I + L L V+LTA+++ +G+ A+ +L GLS FHQLD+
Sbjct: 5 RVALVTGANKGVGFAITRALCRLFSGDVLLTAQDEAQGQAAVQQLQAEGLS-PRFHQLDI 63
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI +L F++ +G L++L+NNA +A K+ +T
Sbjct: 64 TDLQSIRALRDFLRRAYGGLNVLVNNA---------------------VIAFKMEDT--- 99
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIP--NER 179
T + +AE + TN+ G + V LLPL++ + +++ LR P ++
Sbjct: 100 ---TPFHIQAEVTMKTNFDGTRDVCTELLPLMRPGGRVVNVSSMTCLRALKSCSPELQQK 156
Query: 180 LRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK 239
R+E ++E ++ ++KKF+ D K H+ GW AY ++K + +R+ A+
Sbjct: 157 FRSE-----TITEEELVGLMKKFVEDTKKGVHQTEGWPD--TAYGVTKMGVTVLSRIQAR 209
Query: 240 K------NPHMLINCVHPGFVSTDFNWHKGTMTVDEGARGPVMLSLLP--ADGPTG 287
+L+N PG+V TD T + +EGA PV L+LLP A+GP G
Sbjct: 210 HLSEHRGGDKILVNACCPGWVRTDMGGPNATKSPEEGAETPVYLALLPPDAEGPHG 265
>RDHC_HUMAN (Q96NR8) Retinol dehydrogenase 12 (EC 1.1.1.-)
Length = 316
Score = 71.2 bits (173), Expect = 3e-12
Identities = 82/307 (26%), Positives = 123/307 (39%), Gaps = 78/307 (25%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSNVMFHQLDV 61
+ V+TGAN GIG E ++LA G V + R+ +G A +++ T S V+ +LD+
Sbjct: 40 KVVVITGANTGIGKETARELASRGARVYIACRDVLKGESAASEIRVDTKNSQVLVRKLDL 99
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI + A+ + +L ILINNAG K
Sbjct: 100 SDTKSIRAFAEGFLAEEKQLHILINNAGVMMCPYSK------------------------ 135
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
T E L N+ G +T LL L++S A AR+VN+SS+ + +IP
Sbjct: 136 -----TADGFETHLGVNHLGHFLLTYLLLEQLKVS-APARVVNVSSVAHHIGKIP----- 184
Query: 182 NELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK- 240
HD ++ + G+ AY SK + +TR LAK+
Sbjct: 185 ---------------------FHDLQSEKRYSRGF-----AYCHSKLANVLFTRELAKRL 218
Query: 241 -NPHMLINCVHPGFVSTDFNWHKGTM------------TVDEGARGPVMLSLLPADGP-T 286
+ VHPG V ++ H + T EGA+ + +L P +
Sbjct: 219 QGTGVTTYAVHPGVVRSELVRHSSLLCLLWRLFSPFVKTAREGAQTSLHCALAEGLEPLS 278
Query: 287 GCYF-DC 292
G YF DC
Sbjct: 279 GKYFSDC 285
>RDHB_MOUSE (Q9QYF1) Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal
reductase 1) (RalR1) (Prostate short-chain
dehydrogenase/reductase 1) (Androgen-regulated
short-chain dehydrogenase/reductase 1) (Short-chain
aldehyde dehydrogenase) (SCALD) (Cell line MC
Length = 316
Score = 65.5 bits (158), Expect = 2e-10
Identities = 61/182 (33%), Positives = 78/182 (42%), Gaps = 31/182 (17%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQ-TGLSNVMFHQLDV 61
+ A+VTGAN GIG E K LA G V L R+ +G A ++ TG S V +LD+
Sbjct: 39 KVAIVTGANTGIGKETAKDLAQRGARVYLACRDVDKGELAAREIQAVTGNSQVFVRKLDL 98
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI + AK + L +LINNAG K
Sbjct: 99 ADTKSIRAFAKDFLAEEKHLHLLINNAGVMMCPYSK------------------------ 134
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
T E + N+ G +T LL L+ S A +RIVNLSSL L RI L+
Sbjct: 135 -----TADGFEMHIGVNHLGHFLLTHLLLEKLKES-APSRIVNLSSLGHHLGRIHFHNLQ 188
Query: 182 NE 183
E
Sbjct: 189 GE 190
>DHSX_HUMAN (Q8N5I4) Dehydrogenase/reductase SDR family member on
chromosome X precursor (EC 1.1.-.-) (DHRSXY)
Length = 330
Score = 65.1 bits (157), Expect = 2e-10
Identities = 68/267 (25%), Positives = 104/267 (38%), Gaps = 68/267 (25%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSN-VMFHQLDV 61
R A+VTG GIG K LA LG+ V++ ND++ + ++K+ + L++ V F D+
Sbjct: 44 RVAIVTGGTDGIGYSTAKHLARLGMHVIIAGNNDSKAKQVVSKIKEETLNDKVEFLYCDL 103
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
SI + + K L +LINNAG V K
Sbjct: 104 ASMTSIRQFVQKFKMKKIPLHVLINNAGVMMVPQRK------------------------ 139
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPA---KARIVNLSSLRGELKRIPNE 178
T EE NY G +T LL L+ S + AR+V +SS
Sbjct: 140 -----TRDGFEEHFGLNYLGHFLLTNLLLDTLKESGSPGHSARVVTVSS----------- 183
Query: 179 RLRNELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYT---- 234
++ + +D +++ AY+ SK +L +T
Sbjct: 184 --------------------ATHYVAELNMDDLQSSACYSPHAAYAQSKLALVLFTYHLQ 223
Query: 235 RVLAKKNPHMLINCVHPGFVSTDFNWH 261
R+LA + H+ N V PG V+TD H
Sbjct: 224 RLLAAEGSHVTANVVDPGVVNTDLYKH 250
>RDHC_BOVIN (P59837) Retinol dehydrogenase 12 (EC 1.1.1.-) (Double
substrate-specificity short chain
dehydrogenase/reductase 2)
Length = 316
Score = 63.5 bits (153), Expect = 6e-10
Identities = 73/262 (27%), Positives = 104/262 (38%), Gaps = 64/262 (24%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSNVMFHQLDV 61
+ V+TGAN GIG E ++LA G V + R+ +G A +++ T S V+ +LD+
Sbjct: 40 KVVVITGANTGIGKETARELARRGARVYIACRDVLKGESAASEIQADTKNSQVLVRKLDL 99
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI + A+ + +L ILINNAG K
Sbjct: 100 SDTKSIRAFAEGFLAEEKQLHILINNAGVMLCPYSK------------------------ 135
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
T E L N+ G +T LL L+ S A AR+VNLSS+ L +I
Sbjct: 136 -----TADGFETHLAVNHLGHFLLTHLLLGRLKES-APARVVNLSSVAHHLGKIR----- 184
Query: 182 NELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAK-- 239
HD + + + G+ AY SK + +TR LAK
Sbjct: 185 ---------------------FHDLQGDKYYNLGF-----AYCHSKLANVLFTRELAKRL 218
Query: 240 KNPHMLINCVHPGFVSTDFNWH 261
K + VHPG V + H
Sbjct: 219 KGTGVTTYAVHPGIVRSKLVRH 240
>RDHB_HUMAN (Q8TC12) Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal
reductase 1) (RalR1) (Prostate short-chain
dehydrogenase/reductase 1) (Androgen-regulated
short-chain dehydrogenase/reductase 1) (HCV core-binding
protein HCBP12) (CGI-82)
Length = 318
Score = 62.0 bits (149), Expect = 2e-09
Identities = 58/182 (31%), Positives = 77/182 (41%), Gaps = 31/182 (17%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQT-GLSNVMFHQLDV 61
+ VVTGAN GIG E K+LA G V L R+ +G ++ T G V+ +LD+
Sbjct: 42 KVVVVTGANTGIGKETAKELAQRGARVYLACRDVEKGELVAKEIQTTTGNQQVLVRKLDL 101
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI + AK + L +LINNAG K
Sbjct: 102 SDTKSIRAFAKGFLAEEKHLHVLINNAGVMMCPYSK------------------------ 137
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
T E + N+ G +T LL L+ S A +RIVN+SSL L RI L+
Sbjct: 138 -----TADGFEMHIGVNHLGHFLLTHLLLEKLKES-APSRIVNVSSLAHHLGRIHFHNLQ 191
Query: 182 NE 183
E
Sbjct: 192 GE 193
>DHS7_MOUSE (Q9CXR1) Dehydrogenase/reductase SDR family member 7
precursor (EC 1.1.-.-) (Retinal short-chain
dehydrogenase/reductase 4)
Length = 338
Score = 60.5 bits (145), Expect = 5e-09
Identities = 47/167 (28%), Positives = 77/167 (45%), Gaps = 31/167 (18%)
Query: 7 VTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGL---SNVMFHQLDVLD 63
VTGA+ GIG E+ QL+ LGV++VL+AR + + G +++ LD+ D
Sbjct: 55 VTGASSGIGEELAFQLSKLGVSLVLSARRAQELERVKRRCLENGNLKEKDILVLPLDLTD 114
Query: 64 ALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGV 123
S E+ K + +FG++DIL+NN G S + V
Sbjct: 115 TSSHEAATKAVLQEFGKIDILVNNGGRSQRSL---------------------------V 147
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRG 170
L +E +N NY G +T +LP + + + +IV ++S+ G
Sbjct: 148 LETNLDVFKELINLNYIGTVSLTKCVLPHM-IERKQGKIVTVNSIAG 193
>RDHC_MOUSE (Q8BYK4) Retinol dehydrogenase 12 (EC 1.1.1.-)
Length = 316
Score = 60.1 bits (144), Expect = 7e-09
Identities = 69/258 (26%), Positives = 103/258 (39%), Gaps = 64/258 (24%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLH-QTGLSNVMFHQLDV 61
+ V+TGAN GIG E ++LA G V + R+ +G A +++ T S V+ +LD+
Sbjct: 40 KVVVITGANTGIGKETARELARRGARVYIACRDVLKGESAASEIRADTKNSQVLVRKLDL 99
Query: 62 LDALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQ 121
D SI + A+ + +L ILINNAG K
Sbjct: 100 SDTKSIRAFAERFLAEEKKLHILINNAGVMMCPYSK------------------------ 135
Query: 122 GVLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLR 181
T E N+ G +T LL L+ S A AR+VNLSS+ + +I L+
Sbjct: 136 -----TTDGFETHFGVNHLGHFLLTYLLLERLKES-APARVVNLSSIAHLIGKIRFHDLQ 189
Query: 182 NELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK- 240
+ K++ F AY SK + +TR LAK+
Sbjct: 190 GQ----------------KRYCSAF---------------AYGHSKLANLLFTRELAKRL 218
Query: 241 -NPHMLINCVHPGFVSTD 257
+ VHPG V ++
Sbjct: 219 QGTGVTAYAVHPGVVLSE 236
>DHS7_HUMAN (Q9Y394) Dehydrogenase/reductase SDR family member 7
precursor (EC 1.1.-.-) (Retinal short-chain
dehydrogenase/reductase 4) (retSDR4) (CGI-86)
(UNQ285/PRO3448)
Length = 339
Score = 58.5 bits (140), Expect = 2e-08
Identities = 47/167 (28%), Positives = 76/167 (45%), Gaps = 31/167 (18%)
Query: 7 VTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGL---SNVMFHQLDVLD 63
VTGA+ GIG E+ QL+ LGV++VL+AR + + G +++ LD+ D
Sbjct: 55 VTGASSGIGEELAYQLSKLGVSLVLSARRVHELERVKRRCLENGNLKEKDILVLPLDLTD 114
Query: 64 ALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGV 123
S E+ K + +FGR+DIL+NN G S +L
Sbjct: 115 TGSHEAATKAVLQEFGRIDILVNNGGMS------------------------QRSLCMDT 150
Query: 124 LTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRG 170
Y+K + NY G +T +LP + + + +IV ++S+ G
Sbjct: 151 SLDVYRK---LIELNYLGTVSLTKCVLPHM-IERKQGKIVTVNSILG 193
>DHK1_STRVN (P16542) Granaticin polyketide synthase putative
ketoacyl reductase 1 (EC 1.3.1.-) (ORF5)
Length = 272
Score = 58.5 bits (140), Expect = 2e-08
Identities = 49/167 (29%), Positives = 72/167 (42%), Gaps = 27/167 (16%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTGA GIGL I ++LA LG L AR++ R + +L G +V DV D
Sbjct: 20 ALVTGATSGIGLAIARRLAALGARTFLCARDEERLAQTVKELRGEGF-DVDGTVCDVADP 78
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
I + ++G +DIL+NNAG S G + WL +N L
Sbjct: 79 AQIRAYVAAAVQRYGTVDILVNNAGRS------GGGATAEIADELWLDVITTN------L 126
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGE 171
T + +E LN L+ + RI+N++S G+
Sbjct: 127 TSVFLMTKEVLNAGG--------------MLAKKRGRIINIASTGGK 159
>FABG_ECOLI (P25716) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 244
Score = 57.4 bits (137), Expect = 4e-08
Identities = 63/257 (24%), Positives = 109/257 (41%), Gaps = 77/257 (29%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
+ A+VTGA++GIG I + LA G V+ TA ++ G AI+ +M L+V
Sbjct: 6 KIALVTGASRGIGRAIAETLAARGAKVIGTATSEN-GAQAISDYLGANGKGLM---LNVT 61
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
D SIES+ + I+ +FG +DIL+NNAG + ++ L + + W
Sbjct: 62 DPASIESVLEKIRAEFGEVDILVNNAG-----ITRDNL-LMRMKDEEW------------ 103
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQLSPAKARIVNLSSLRGELKRIPNERLRN 182
+ + TN V R++ A++ + + RI+ + S+ G +
Sbjct: 104 ---------NDIIETNLSSVFRLSKAVMRAM-MKKRHGRIITIGSVVGTM---------- 143
Query: 183 ELGDVDELSEGKIDAMVKKFLHDFKANDHEANGWGMMLPAYSISKASLNAYTRVLAKK-- 240
N +AN Y+ +KA L +++ LA++
Sbjct: 144 -------------------------GNGGQAN--------YAAAKAGLIGFSKSLAREVA 170
Query: 241 NPHMLINCVHPGFVSTD 257
+ + +N V PGF+ TD
Sbjct: 171 SRGITVNVVAPGFIETD 187
>DHKR_STRCM (P41177) Monensin polyketide synthase putative ketoacyl
reductase (EC 1.3.1.-) (ORF5)
Length = 261
Score = 57.0 bits (136), Expect = 6e-08
Identities = 50/170 (29%), Positives = 78/170 (45%), Gaps = 29/170 (17%)
Query: 3 RYAVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVL 62
R A+VTGA GIGL + LA G V L AR ++ + L GL LDV
Sbjct: 7 RVALVTGATSGIGLATARLLAAQGHLVFLGARTESDVIATVKALRNDGLE-AEGQVLDVR 65
Query: 63 DALSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQG 122
D S+ + + ++GR+D+L+NNAG S G V+ L
Sbjct: 66 DGASVTAFVQAAVDRYGRIDVLVNNAGRS--------------------GGGVTADL--- 102
Query: 123 VLTQTYKKAEECLNTNYYGVKRVTMALLPLLQL-SPAKARIVNLSSLRGE 171
T + ++ ++TN V R+T A+L + + + RI+N++S G+
Sbjct: 103 ----TDELWDDVIDTNLNSVFRMTRAVLTTGGMRTRERGRIINVASTAGK 148
>ACT3_STRCO (P16544) Putative ketoacyl reductase (EC 1.3.1.-)
Length = 261
Score = 57.0 bits (136), Expect = 6e-08
Identities = 49/168 (29%), Positives = 73/168 (43%), Gaps = 29/168 (17%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDA 64
A+VTGA GIGLEI ++L G+ V + AR + R + +L + G+ DV
Sbjct: 9 ALVTGATSGIGLEIARRLGKEGLRVFVCARGEEGLRTTLKELREAGV-EADGRTCDVRSV 67
Query: 65 LSIESLAKFIQHKFGRLDILINNAGASCVEVDKEGLKALNVDPATWLAGKVSNTLLQGVL 124
IE+L + ++G +D+L+NNAG G + WL
Sbjct: 68 PEIEALVAAVVERYGPVDVLVNNAGR------PGGGATAELADELWL------------- 108
Query: 125 TQTYKKAEECLNTNYYGVKRVTMALLPL-LQLSPAKARIVNLSSLRGE 171
+ + TN GV RVT +L L RIVN++S G+
Sbjct: 109 --------DVVETNLTGVFRVTKQVLKAGGMLERGTGRIVNIASTGGK 148
>BDHA_ALCEU (Q9X6U2) D-beta-hydroxybutyrate dehydrogenase (EC
1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase)
(3-HBDH)
Length = 258
Score = 55.8 bits (133), Expect = 1e-07
Identities = 33/86 (38%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query: 5 AVVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLS-NVMFHQLDVLD 63
A+VTG+ GIGL I K LA G +++ D A ++ Q G V +H D+
Sbjct: 7 ALVTGSTSGIGLGIAKALAAQGANIIVNGFGDADAAKA--EIAQAGQGIRVGYHGADMSK 64
Query: 64 ALSIESLAKFIQHKFGRLDILINNAG 89
A IE + ++ Q FG DIL+NNAG
Sbjct: 65 AAEIEDMMRYAQSDFGGADILVNNAG 90
>DLTE_BACSU (P39577) Protein dltE
Length = 252
Score = 55.5 bits (132), Expect = 2e-07
Identities = 36/102 (35%), Positives = 59/102 (57%), Gaps = 7/102 (6%)
Query: 6 VVTGANKGIGLEIVKQLAFLGVTVVLTARNDTRGRDAITKLHQTGLSNVMFHQLDVLDAL 65
++TG + GIGLE+ K+L LG V++ R++ R +A + L N+ Q DV D
Sbjct: 9 LITGGSAGIGLELAKRLLELGNEVIICGRSEARLAEA-----KQQLPNIHTKQCDVADRS 63
Query: 66 SIESLAKFIQHKFGRLDILINNAGASCVEVD-KEGLKALNVD 106
E+L ++ ++ L++L+NNAG E+D K+G + L VD
Sbjct: 64 QREALYEWALKEYPNLNVLVNNAGIQ-KEIDFKKGTEELFVD 104
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,452,690
Number of Sequences: 164201
Number of extensions: 1338856
Number of successful extensions: 3859
Number of sequences better than 10.0: 297
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 3467
Number of HSP's gapped (non-prelim): 376
length of query: 298
length of database: 59,974,054
effective HSP length: 109
effective length of query: 189
effective length of database: 42,076,145
effective search space: 7952391405
effective search space used: 7952391405
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146743.8


