

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146723.15 + phase: 0
(345 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
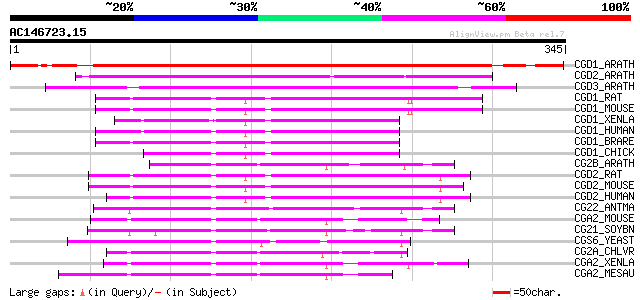
Score E
Sequences producing significant alignments: (bits) Value
CGD1_ARATH (P42751) Cyclin delta-1 358 1e-98
CGD2_ARATH (P42752) Cyclin delta-2 182 1e-45
CGD3_ARATH (P42753) Cyclin delta-3 172 1e-42
CGD1_RAT (P39948) G1/S-specific cyclin D1 83 1e-15
CGD1_MOUSE (P25322) G1/S-specific cyclin D1 81 3e-15
CGD1_XENLA (P50755) G1/S-specific cyclin D1 80 8e-15
CGD1_HUMAN (P24385) G1/S-specific cyclin D1 (PRAD1 oncogene) (BC... 80 1e-14
CGD1_BRARE (Q90459) G1/S-specific cyclin D1 80 1e-14
CGD1_CHICK (P55169) G1/S-specific cyclin D1 78 4e-14
CG2B_ARATH (P30183) G2/mitotic-specific cyclin (B-like cyclin) 78 4e-14
CGD2_RAT (Q04827) G1/S-specific cyclin D2 (Vin-1 proto-oncogene) 77 5e-14
CGD2_MOUSE (P30280) G1/S-specific cyclin D2 77 6e-14
CGD2_HUMAN (P30279) G1/S-specific cyclin D2 77 8e-14
CG22_ANTMA (P34801) G2/mitotic-specific cyclin 2 77 8e-14
CGA2_MOUSE (P51943) Cyclin A2 (Cyclin A) 75 3e-13
CG21_SOYBN (P25011) G2/mitotic-specific cyclin S13-6 (B-like cyc... 75 3e-13
CGS6_YEAST (P32943) S-phase entry cyclin 6 74 5e-13
CG2A_CHLVR (P51986) G2/mitotic-specific cyclin A (Fragment) 74 7e-13
CGA2_XENLA (P47827) Cyclin A2 72 2e-12
CGA2_MESAU (P37881) Cyclin A2 (Cyclin A) 72 2e-12
>CGD1_ARATH (P42751) Cyclin delta-1
Length = 335
Score = 358 bits (918), Expect = 1e-98
Identities = 198/347 (57%), Positives = 247/347 (71%), Gaps = 27/347 (7%)
Query: 1 MPLSSTSDCELLCGEDSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEH 60
M +S ++D +L CGEDS V +G E + D SS P +SIA FIE
Sbjct: 12 MSVSFSNDMDLFCGEDSG-VFSG---ESTVDFSSSEVDSWPG---------DSIACFIED 58
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E FVPG DY+SRFQ+RSL++S RE+++AWILKV YY FQPLTAYL+VNYMDRFL +R
Sbjct: 59 ERHFVPGHDYLSRFQTRSLDASAREDSVAWILKVQAYYNFQPLTAYLAVNYMDRFLYARR 118
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
LPE++GWP+QLL+VACLSLAAKMEE LVPSL DFQ+ G KY+F+ +TI RMELLVL++LD
Sbjct: 119 LPETSGWPMQLLAVACLSLAAKMEEILVPSLFDFQVAGVKYLFEAKTIKRMELLVLSVLD 178
Query: 181 WRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAIL 240
WRLRS+TP F+SFFA K+D +GTF F IS ATEIILSNI++ASFL Y PS IAAAAIL
Sbjct: 179 WRLRSVTPFDFISFFAYKIDPSGTFLGFFISHATEIILSNIKEASFLEYWPSSIAAAAIL 238
Query: 241 SAANEIPNW-SFVNP-EHAESWCEGLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVT 298
ANE+P+ S VNP E E+WC+GLSKEKI+ CY L++ + NN+ N PKV+ +LRV+
Sbjct: 239 CVANELPSLSSVVNPHESPETWCDGLSKEKIVRCYRLMKAMAIENNRLNTPKVIAKLRVS 298
Query: 299 ARTRRWSTVSSSSSSPS-SSSSPSFSLSYKKRKLNSCFWVDVDKGNS 344
R SS+ + PS SSSP K+RKL+ WV + S
Sbjct: 299 VR------ASSTLTRPSDESSSPC-----KRRKLSGYSWVGDETSTS 334
>CGD2_ARATH (P42752) Cyclin delta-2
Length = 361
Score = 182 bits (462), Expect = 1e-45
Identities = 106/259 (40%), Positives = 153/259 (58%), Gaps = 7/259 (2%)
Query: 42 SSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQ 101
SSSL E+ I + E +F PG DYV R S L+ S R +A+ WILKV +Y F
Sbjct: 59 SSSL----SEDRIKEMLVREIEFCPGTDYVKRLLSGDLDLSVRNQALDWILKVCAHYHFG 114
Query: 102 PLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKY 161
L LS+NY+DRFL S LP+ W QLL+V+CLSLA+KMEE VP ++D Q+E K+
Sbjct: 115 HLCICLSMNYLDRFLTSYELPKDKDWAAQLLAVSCLSLASKMEETDVPHIVDLQVEDPKF 174
Query: 162 IFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNI 221
+F+ +TI RMELLV+T L+WRL+++TP SF+ +F K+ +G + +I R++ IL+
Sbjct: 175 VFEAKTIKRMELLVVTTLNWRLQALTPFSFIDYFVDKI--SGHVSENLIYRSSRFILNTT 232
Query: 222 QDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVS 281
+ FL +RPS IAAAA +S + ++ E A S + +E++ C L++ +
Sbjct: 233 KAIEFLDFRPSEIAAAAAVSVSIS-GETECIDEEKALSSLIYVKQERVKRCLNLMRSLTG 291
Query: 282 SNNQRNAPKVLPQLRVTAR 300
N R Q RV R
Sbjct: 292 EENVRGTSLSQEQARVAVR 310
>CGD3_ARATH (P42753) Cyclin delta-3
Length = 376
Score = 172 bits (436), Expect = 1e-42
Identities = 105/294 (35%), Positives = 169/294 (56%), Gaps = 15/294 (5%)
Query: 23 GDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSRSLESS 82
G+ E +S L SSSS + E+E+ + +F + E + + D V S+
Sbjct: 33 GEEVEENSSLSSSSSPFVVLQQDLFWEDEDLVTLFSKEEEQGLSCLDDVYL-------ST 85
Query: 83 TREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAK 142
R+EA+ WIL+V+ +YGF L A L++ Y+D+F+ S L W LQL+SVACLSLAAK
Sbjct: 86 DRKEAVGWILRVNAHYGFSTLAAVLAITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAK 145
Query: 143 MEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDST 202
+EE VP LLDFQ+E KY+F+ +TI RMELL+L+ L+W++ ITP+SF+ +L
Sbjct: 146 VEETQVPLLLDFQVEETKYVFEAKTIQRMELLILSTLEWKMHLITPISFVDHIIRRLGLK 205
Query: 203 GTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCE 262
+++ ++LS I D+ F+ Y PS +AAA ++ ++ + ++ +
Sbjct: 206 NNAHWDFLNKCHRLLLSVISDSRFVGYLPSVVAAATMMRIIEQVDPFDPLSYQTNLLGVL 265
Query: 263 GLSKEKIIGCYELIQEIVSSNNQRNAPKVLPQLRVTARTRRWSTVSSSS-SSPS 315
L+KEK+ CY+LI ++ ++ Q+++ + +R S SSSS +SPS
Sbjct: 266 NLTKEKVKTCYDLILQL-------PVDRICLQIQIQSSKKRKSHDSSSSLNSPS 312
>CGD1_RAT (P39948) G1/S-specific cyclin D1
Length = 295
Score = 82.8 bits (203), Expect = 1e-15
Identities = 66/249 (26%), Positives = 112/249 (44%), Gaps = 15/249 (6%)
Query: 54 IAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMD 113
+ ++ E P Y Q R + S R+ W+L+V E + L++NY+D
Sbjct: 28 LRAMLKTEETCAPSVSYFKCVQ-REIVPSMRKIVATWMLEVCEEQKCEEEVFPLAMNYLD 86
Query: 114 RFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRM 171
RFL PL +S LQLL C+ +A+KM+E PL L + + +P +L+M
Sbjct: 87 RFLSLEPLKKSR---LQLLGATCMFVASKMKETIPLTAEKLCIYTDNS---IRPEELLQM 140
Query: 172 ELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRP 231
ELL++ L W L ++TP F+ F K+ I A + D F++ P
Sbjct: 141 ELLLVNKLKWNLAAMTPHDFIEHFLSKMPEADENKQIIRKHAQTFVALCATDVKFISNPP 200
Query: 232 SCIAAAAILSAANEI----PN--WSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQ 285
S +AA ++++A + PN S H S + + C E I+ ++ S+ +
Sbjct: 201 SMVAAGSVVAAMQGLNLGSPNNFLSCYRTTHFLSRVIKCDPDCLRACQEQIEALLESSLR 260
Query: 286 RNAPKVLPQ 294
+ + P+
Sbjct: 261 QAQQNIDPK 269
>CGD1_MOUSE (P25322) G1/S-specific cyclin D1
Length = 295
Score = 81.3 bits (199), Expect = 3e-15
Identities = 66/249 (26%), Positives = 112/249 (44%), Gaps = 15/249 (6%)
Query: 54 IAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMD 113
+ ++ E P Y Q + + S R+ W+L+V E + L++NY+D
Sbjct: 28 LRAMLKTEETCAPSVSYFKCVQ-KEIVPSMRKIVATWMLEVCEEQKCEEEVFPLAMNYLD 86
Query: 114 RFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRM 171
RFL PL +S LQLL C+ +A+KM+E PL L + + +P +L+M
Sbjct: 87 RFLSLEPLKKSR---LQLLGATCMFVASKMKETIPLTAEKLCIYTDNS---IRPEELLQM 140
Query: 172 ELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRP 231
ELL++ L W L ++TP F+ F K+ I A + D F++ P
Sbjct: 141 ELLLVNKLKWNLAAMTPHDFIEHFLSKMPEADENKQTIRKHAQTFVALCATDVKFISNPP 200
Query: 232 SCIAAAAILSAANEI----PN--WSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQ 285
S +AA ++++A + PN S H S + + C E I+ ++ S+ +
Sbjct: 201 SMVAAGSVVAAMQGLNLGSPNNFLSCYRTTHFLSRVIKCDPDCLRACQEQIEALLESSLR 260
Query: 286 RNAPKVLPQ 294
+ V P+
Sbjct: 261 QAQQNVDPK 269
>CGD1_XENLA (P50755) G1/S-specific cyclin D1
Length = 291
Score = 80.1 bits (196), Expect = 8e-15
Identities = 60/179 (33%), Positives = 86/179 (47%), Gaps = 9/179 (5%)
Query: 66 PGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESN 125
P Y Q L + R+ W+L+V E + L++NY+DRFL PL +S
Sbjct: 38 PSMSYFKCVQKEILPNM-RKIVATWMLEVCEEQKCEEEVFPLAMNYLDRFLSVEPLRKS- 95
Query: 126 GWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRL 183
W LQLL C+ LA+KM+E PL L + + +P +L MEL VL L W L
Sbjct: 96 -W-LQLLGATCMFLASKMKETIPLTAEKLCIYTDNS---IRPDELLIMELRVLNKLKWDL 150
Query: 184 RSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
S+TP F+ F K+ T I A + D +F++ PS IAA ++ +A
Sbjct: 151 ASVTPHDFIEHFLNKMPLTEDTKQIIRKHAQTFVALCATDVNFISNPPSMIAAGSVAAA 209
>CGD1_HUMAN (P24385) G1/S-specific cyclin D1 (PRAD1 oncogene) (BCL-1
oncogene)
Length = 295
Score = 79.7 bits (195), Expect = 1e-14
Identities = 55/191 (28%), Positives = 90/191 (46%), Gaps = 9/191 (4%)
Query: 54 IAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMD 113
+ ++ E P Y Q L S R+ W+L+V E + L++NY+D
Sbjct: 28 LRAMLKAEETCAPSVSYFKCVQKEVLPSM-RKIVATWMLEVCEEQKCEEEVFPLAMNYLD 86
Query: 114 RFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRM 171
RFL P+ +S LQLL C+ +A+KM+E PL L + + +P +L+M
Sbjct: 87 RFLSLEPVKKSR---LQLLGATCMFVASKMKETIPLTAEKLCIYTDNS---IRPEELLQM 140
Query: 172 ELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRP 231
ELL++ L W L ++TP F+ F K+ I A + D F++ P
Sbjct: 141 ELLLVNKLKWNLAAMTPHDFIEHFLSKMPEAEENKQIIRKHAQTFVALCATDVKFISNPP 200
Query: 232 SCIAAAAILSA 242
S +AA ++++A
Sbjct: 201 SMVAAGSVVAA 211
>CGD1_BRARE (Q90459) G1/S-specific cyclin D1
Length = 291
Score = 79.7 bits (195), Expect = 1e-14
Identities = 56/191 (29%), Positives = 91/191 (47%), Gaps = 9/191 (4%)
Query: 54 IAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMD 113
+ ++ E ++P +Y Q + + R+ W+L+V E + L++NY+D
Sbjct: 28 LQTMLKAEENYLPSPNYFKCVQ-KEIVPKMRKIVATWMLEVCEEQKCEEEVFPLAMNYLD 86
Query: 114 RFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRM 171
RFL P ++ LQLL C+ LA+KM+E PL L + + +P +L+M
Sbjct: 87 RFLSVEPTKKTR---LQLLGATCMFLASKMKETVPLTAEKLCIYTDNS---VRPGELLQM 140
Query: 172 ELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRP 231
ELL L L W L S+TP F+ F KL + + A + D +F+ P
Sbjct: 141 ELLALNKLKWDLASVTPHDFIEHFLAKLPIHQSSKQILRKHAQTFVALCATDVNFIASPP 200
Query: 232 SCIAAAAILSA 242
S IAA ++ +A
Sbjct: 201 SMIAAGSVAAA 211
>CGD1_CHICK (P55169) G1/S-specific cyclin D1
Length = 292
Score = 77.8 bits (190), Expect = 4e-14
Identities = 51/161 (31%), Positives = 81/161 (49%), Gaps = 8/161 (4%)
Query: 84 REEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKM 143
R+ W+L+V E + L++NY+DRFL PL +S LQLL C+ +A+KM
Sbjct: 57 RKIVATWMLEVCEEQKCEEEVFPLAMNYLDRFLSFEPLKKSR---LQLLGATCMFVASKM 113
Query: 144 EE--PLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDS 201
+E PL L + + +P +L+MELL++ L W L ++TP F+ F K+
Sbjct: 114 KETIPLTAEKLCIYTDNS---IRPDELLQMELLLVNKLKWNLAAMTPHDFIEHFLTKMPL 170
Query: 202 TGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSA 242
I A + D F++ PS IAA ++++A
Sbjct: 171 AEDTKQIIRKHAQTFVALCATDVKFISNPPSMIAAGSVVAA 211
>CG2B_ARATH (P30183) G2/mitotic-specific cyclin (B-like cyclin)
Length = 428
Score = 77.8 bits (190), Expect = 4e-14
Identities = 54/195 (27%), Positives = 98/195 (49%), Gaps = 21/195 (10%)
Query: 88 IAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPL 147
+ W++ VH + P T YL+VN +DRFL +P+P LQL+ ++ L ++AK EE
Sbjct: 201 VEWLIDVHVRFELNPETFYLTVNILDRFLSVKPVPRKE---LQLVGLSALLMSAKYEEIW 257
Query: 148 VPSLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFF---ACKLDSTGT 204
P + D ++ A + + + IL ME +L+ L+W L T FL+ F + +
Sbjct: 258 PPQVEDL-VDIADHAYSHKQILVMEKTILSTLEWYLTVPTHYVFLARFIKASIADEKMEN 316
Query: 205 FTHFIISRATEIILSNIQDASFLTYRPSCIAAAAILSAAN---EIPNWSFVNPEHAESWC 261
H++ L + + + + PS +AA+AI +A + ++P W+ H
Sbjct: 317 MVHYLAE------LGVMHYDTMIMFSPSMVAASAIYAARSSLRQVPIWTSTLKHHT---- 366
Query: 262 EGLSKEKIIGCYELI 276
G S+ +++ C +L+
Sbjct: 367 -GYSETQLMDCAKLL 380
>CGD2_RAT (Q04827) G1/S-specific cyclin D2 (Vin-1 proto-oncogene)
Length = 288
Score = 77.4 bits (189), Expect = 5e-14
Identities = 65/245 (26%), Positives = 111/245 (44%), Gaps = 15/245 (6%)
Query: 50 EEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSV 109
E+ + + E +++P Y Q + ++ R W+L+V E + L++
Sbjct: 22 EDRVLQNLLTIEERYLPQCSYFKCVQ-KDIQPYMRRMVATWMLEVCEEQKCEEEVFPLAM 80
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRT 167
NY+DRFL P P+++ LQLL C+ LA+K++E PL L + + +P+
Sbjct: 81 NYLDRFLAGVPTPKTH---LQLLGAVCMFLASKLKETIPLTAEKLCIYTDNS---VKPQE 134
Query: 168 ILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFL 227
+L EL+VL L W L ++TP F+ KL I A I D F
Sbjct: 135 LLEWELVVLGKLKWNLAAVTPHDFIEHILRKLPQQKEKLSLIRKHAQTFIALCATDFKFA 194
Query: 228 TYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSK------EKIIGCYELIQEIVS 281
Y PS IA ++ +A + VN ++ E L+K + + C E I+ ++
Sbjct: 195 MYPPSMIATGSVGAAICGLQQDEEVNALTCDALTELLTKITHTDVDCLKACQEQIEAVLL 254
Query: 282 SNNQR 286
++ Q+
Sbjct: 255 NSLQQ 259
>CGD2_MOUSE (P30280) G1/S-specific cyclin D2
Length = 289
Score = 77.0 bits (188), Expect = 6e-14
Identities = 64/237 (27%), Positives = 106/237 (44%), Gaps = 11/237 (4%)
Query: 50 EEESIAVFIEHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSV 109
E+ + + E +++P Y Q + ++ R W+L+V E + L++
Sbjct: 22 EDRVLQNLLTIEERYLPQCSYFKCVQ-KDIQPYMRRMVATWMLEVCEEQKCEEEVFPLAM 80
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRT 167
NY+DRFL P P+++ LQLL C+ LA+K++E PL L + + +P+
Sbjct: 81 NYLDRFLAGVPTPKTH---LQLLGAVCMFLASKLKETIPLTAEKLCIYTDNS---VKPQE 134
Query: 168 ILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFL 227
+L EL+VL L W L ++TP F+ KL I A I D F
Sbjct: 135 LLEWELVVLGKLKWNLAAVTPHDFIEHILRKLPQQKEKLSLIRKHAQTFIALCATDFKFA 194
Query: 228 TYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSK--EKIIGCYELIQEIVSS 282
Y PS IA ++ +A + VN ++ E L+K + C + QE + +
Sbjct: 195 MYPPSMIATGSVGAAICGLQQDDEVNTLTCDALTELLAKITHTDVDCLKACQEQIEA 251
>CGD2_HUMAN (P30279) G1/S-specific cyclin D2
Length = 289
Score = 76.6 bits (187), Expect = 8e-14
Identities = 64/234 (27%), Positives = 107/234 (45%), Gaps = 15/234 (6%)
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E +++P Y Q + ++ R W+L+V E + L++NY+DRFL P
Sbjct: 34 EERYLPQCSYFKCVQ-KDIQPYMRRMVATWMLEVCEEQKCEEEVFPLAMNYLDRFLAGVP 92
Query: 121 LPESNGWPLQLLSVACLSLAAKMEE--PLVPSLLDFQIEGAKYIFQPRTILRMELLVLTI 178
P+S+ LQLL C+ LA+K++E PL L + + +P+ +L EL+VL
Sbjct: 93 TPKSH---LQLLGAVCMFLASKLKETSPLTAEKLCIYTDNS---IKPQELLEWELVVLGK 146
Query: 179 LDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAA 238
L W L ++TP F+ KL I A I D F Y PS IA +
Sbjct: 147 LKWNLAAVTPHDFIEHILRKLPQQREKLSLIRKHAQTFIALCATDFKFAMYPPSMIATGS 206
Query: 239 ILSAANEIPNWSFVNPEHAESWCEGLSK------EKIIGCYELIQEIVSSNNQR 286
+ +A + V+ ++ E L+K + + C E I+ ++ ++ Q+
Sbjct: 207 VGAAICGLQQDEEVSSLTCDALTELLAKITNTDVDCLKACQEQIEAVLLNSLQQ 260
>CG22_ANTMA (P34801) G2/mitotic-specific cyclin 2
Length = 441
Score = 76.6 bits (187), Expect = 8e-14
Identities = 65/234 (27%), Positives = 107/234 (44%), Gaps = 23/234 (9%)
Query: 53 SIAVFIEHEFKFVPGFDYVSR-----FQSRSLESSTREEAIAWILKVHEYYGFQPLTAYL 107
++ ++E +KF + SR + R I W+++VH + P T YL
Sbjct: 185 AVVEYVEDMYKFYKSAENDSRPHDYMDSQPEINEKMRAILIDWLVQVHYKFELSPETLYL 244
Query: 108 SVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQ-IEGAKYIFQPR 166
++N +DR+L S+ LQLL ++ + +A+K EE P + D I Y
Sbjct: 245 TINIVDRYLASKTTSRRE---LQLLGMSSMLIASKYEEIWAPEVNDLVCISDGSY--SNE 299
Query: 167 TILRMELLVLTILDWRLRSITPLSFL-SFFACKLDSTGTFTHFIISRATEIILSNIQDAS 225
+LRME +L L+W L TP FL F L + + + A L + A+
Sbjct: 300 QVLRMEKKILGALEWYLTVPTPYVFLVRFIKASLPDSDVEKNMVYFLAE---LGMMNYAT 356
Query: 226 FLTYRPSCIAAAAILSA---ANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELI 276
+ Y PS IAAAA+ +A N++P W+ H G S+ +++ C +L+
Sbjct: 357 IIMYCPSMIAAAAVYAARCTLNKMPIWNETLRMHT-----GFSEVQLMDCAKLL 405
>CGA2_MOUSE (P51943) Cyclin A2 (Cyclin A)
Length = 422
Score = 74.7 bits (182), Expect = 3e-13
Identities = 68/225 (30%), Positives = 104/225 (46%), Gaps = 32/225 (14%)
Query: 51 EESIAVFI-EHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSV 109
+E I ++ E E K P Y+ R + +S R + W+++V E Y Q T +L+V
Sbjct: 169 QEDIHTYLREMEVKCKPKVGYMKR--QPDITNSMRAILVDWLVEVGEEYKLQNETLHLAV 226
Query: 110 NYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTIL 169
NY+DRFL S + LQL+ A + LA+K EE P + +F + + + +L
Sbjct: 227 NYIDRFLSSMSVLRGK---LQLVGTAAMLLASKFEEIYPPEVAEF-VYITDDTYSKKQVL 282
Query: 170 RMELLVLTILDWRLRSITPLSFLSFF-------ACKLDSTGTFTHFIISRATEIILSNIQ 222
RME LVL +L + L + T FL+ + CK++S F LS I
Sbjct: 283 RMEHLVLKVLAFDLAAPTVNQFLTQYFLHLQPANCKVESLAMFLG---------ELSLID 333
Query: 223 DASFLTYRPSCIAAAAILSAANEIPNWSFVNPEHAESWCEGLSKE 267
+L Y PS IA AA A + +SW E L+++
Sbjct: 334 ADPYLKYLPSLIAGAAFHLALYTVT---------GQSWPESLAQQ 369
>CG21_SOYBN (P25011) G2/mitotic-specific cyclin S13-6 (B-like
cyclin)
Length = 454
Score = 74.7 bits (182), Expect = 3e-13
Identities = 68/239 (28%), Positives = 112/239 (46%), Gaps = 27/239 (11%)
Query: 49 EEEESIAVFIEHEFKFVPGFDYVSR---FQSRSLESSTREEAIA--WILKVHEYYGFQPL 103
+ E + +I+ +KF + SR + E + R AI W++ VH +
Sbjct: 186 DNELAAVEYIDDIYKFYKLVENESRPHDYIGSQPEINERMRAILVDWLIDVHTKFELSLE 245
Query: 104 TAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIF 163
T YL++N +DRFL + +P LQL+ ++ + +A+K EE P + DF + +
Sbjct: 246 TLYLTINIIDRFLAVKTVPRRE---LQLVGISAMLMASKYEEIWPPEVNDFVCLSDR-AY 301
Query: 164 QPRTILRMELLVLTILDWRLRSITPLSFLSFF---ACKLDSTGTFTHFIISRATEIILSN 220
IL ME +L L+W L TPL FL F + HF+ +E+ + N
Sbjct: 302 THEHILTMEKTILNKLEWTLTVPTPLVFLVRFIKASVPDQELDNMAHFL----SELGMMN 357
Query: 221 IQDASFLTYRPSCIAAAAILSA---ANEIPNWSFVNPEHAESWCEGLSKEKIIGCYELI 276
+ L Y PS +AA+A+L+A N+ P W+ H G S+E+++ C L+
Sbjct: 358 Y---ATLMYCPSMVAASAVLAARCTLNKAPFWNETLKLHT-----GYSQEQLMDCARLL 408
>CGS6_YEAST (P32943) S-phase entry cyclin 6
Length = 380
Score = 73.9 bits (180), Expect = 5e-13
Identities = 61/219 (27%), Positives = 105/219 (47%), Gaps = 16/219 (7%)
Query: 37 SSQLPSSSLFAEEEEESIAVFIEHEFKFVPGFDYVSRFQSR-SLESSTREEAIAWILKVH 95
S ++ + AE + + E E + +P +Y+ QS L+SS R I W+++VH
Sbjct: 108 SIEMDDPFMVAEYTDSIFSHLYEKEIQMLPTHNYLMDTQSPYHLKSSMRALLIDWLVEVH 167
Query: 96 EYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQ 155
E + P T +L++N +DRFL + + LQLL + CL +A K EE +P + +F
Sbjct: 168 EKFHCLPETLFLAINLLDRFLSQNVVKLNK---LQLLCITCLFIACKFEEVKLPKITNFA 224
Query: 156 --IEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRA 213
+GA + I + EL VL+ L + + PL+F+ ++ + + A
Sbjct: 225 YVTDGAATV---EGIRKAELFVLSSLGYNISLPNPLNFIR----RISKADNYCIETRNMA 277
Query: 214 TEIILSNIQDASFLTYRPSCIAAAAILSA---ANEIPNW 249
I+ +I F+ +PS +AA ++ A NE W
Sbjct: 278 KFIMEYSICCNKFIHLKPSYLAAMSMYIARKIKNENSKW 316
>CG2A_CHLVR (P51986) G2/mitotic-specific cyclin A (Fragment)
Length = 420
Score = 73.6 bits (179), Expect = 7e-13
Identities = 63/192 (32%), Positives = 93/192 (47%), Gaps = 14/192 (7%)
Query: 61 EFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDSRP 120
E K+ P +Y+ + + SS R I W+++V E Y P T YLSV+Y+DRFL
Sbjct: 177 EAKYRPKSNYMRK--QTDINSSMRAILIDWLVEVSEEYKLIPQTLYLSVSYIDRFLSHMS 234
Query: 121 LPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTILD 180
+ LQL+ AC+ +AAK EE P + +F + + + +LRME L+L L
Sbjct: 235 VLRGK---LQLVGAACMLVAAKFEEIYPPEVAEF-VYITDDTYTAKQVLRMEHLILKTLA 290
Query: 181 WRLRSITPLSFLS--FFACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAA 238
+ L T FLS FA ++ +E+ L N + + Y PS IAA++
Sbjct: 291 FDLSVPTCRDFLSRYLFAANAKPESQL-KYLAEYLSELTLINCDIS--VKYAPSMIAASS 347
Query: 239 ILSA---ANEIP 247
I A N IP
Sbjct: 348 ICVANHMLNSIP 359
>CGA2_XENLA (P47827) Cyclin A2
Length = 415
Score = 72.4 bits (176), Expect = 2e-12
Identities = 70/235 (29%), Positives = 106/235 (44%), Gaps = 24/235 (10%)
Query: 59 EHEFKFVPGFDYVSRFQSRSLESSTREEAIAWILKVHEYYGFQPLTAYLSVNYMDRFLDS 118
E E K P Y+ + + + R + W+++V E Y Q T YL+VNY+DRFL S
Sbjct: 170 EMEVKCKPKAGYMQK--QPDITGNMRAILVDWLVEVGEEYKLQNETLYLAVNYIDRFLSS 227
Query: 119 RPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRMELLVLTI 178
+ LQL+ A + LA+K EE P + +F + + + +L+ME LVL +
Sbjct: 228 MSVLRGK---LQLVGTAAMLLASKFEEIYPPEVAEF-VYITDDTYTKKQVLKMEHLVLKV 283
Query: 179 LDWRLRSITPLSFLSFF------ACKLDSTGTFTHFIISRATEIILSNIQDASFLTYRPS 232
L + L + T L +L+ + + K++S F LS + FL Y PS
Sbjct: 284 LSFDLAAPTILQYLNQYFQIHPVSPKVESLSMFLG---------ELSLVDADPFLRYLPS 334
Query: 233 CIAAAAILSAANEI--PNWSFVNPEHAESWCEGLSKEKIIGCYELIQEIVSSNNQ 285
+AAAA + A I WS E+ E L K I+ Y+ S Q
Sbjct: 335 VVAAAAFVIANCTINERTWSDPLVEYTSYTLETL-KPCILDLYQTYLSAASHQQQ 388
>CGA2_MESAU (P37881) Cyclin A2 (Cyclin A)
Length = 421
Score = 72.4 bits (176), Expect = 2e-12
Identities = 65/216 (30%), Positives = 99/216 (45%), Gaps = 23/216 (10%)
Query: 31 DLDSSSSSQLPSSSLFAEEEEESIAVFI-EHEFKFVPGFDYVSRFQSRSLESSTREEAIA 89
D+ + P S + E I ++ E E K P Y+ + + +S R +
Sbjct: 148 DMSIVLEEEKPVSVNEVPDYHEDIHTYLREMEIKCKPKVGYMKK--QPDITNSMRAILVD 205
Query: 90 WILKVHEYYGFQPLTAYLSVNYMDRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVP 149
W+++V E Y Q T +L+VNY+DRFL S + LQL+ A + LA+K EE P
Sbjct: 206 WLVEVGEEYKLQNETLHLAVNYIDRFLSSMSVLRGK---LQLVGTAAMLLASKFEEIYPP 262
Query: 150 SLLDFQIEGAKYIFQPRTILRMELLVLTILDWRLRSITPLSFLSFF-------ACKLDST 202
+ +F + + + +LRME LVL +L + L + T FL+ + CK++S
Sbjct: 263 EVAEF-VYITDDTYSKKQVLRMEHLVLKVLAFDLAAPTVNQFLNQYFLHQQPANCKVESL 321
Query: 203 GTFTHFIISRATEIILSNIQDASFLTYRPSCIAAAA 238
F LS I +L Y PS IA AA
Sbjct: 322 AMFLG---------ELSLIDADPYLKYLPSLIAGAA 348
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,125,136
Number of Sequences: 164201
Number of extensions: 1572615
Number of successful extensions: 7717
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 59
Number of HSP's that attempted gapping in prelim test: 7321
Number of HSP's gapped (non-prelim): 210
length of query: 345
length of database: 59,974,054
effective HSP length: 111
effective length of query: 234
effective length of database: 41,747,743
effective search space: 9768971862
effective search space used: 9768971862
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146723.15


