

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146719.10 + phase: 0 /pseudo
(112 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
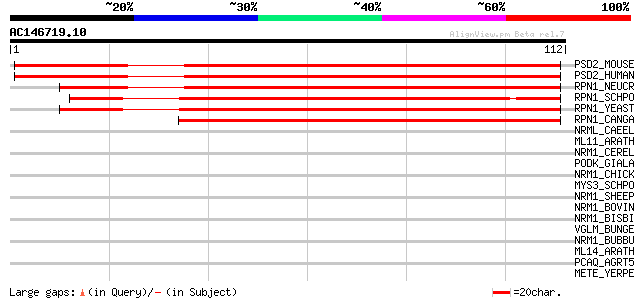
Score E
Sequences producing significant alignments: (bits) Value
PSD2_MOUSE (Q8VDM4) 26S proteasome non-ATPase regulatory subunit... 123 7e-29
PSD2_HUMAN (Q13200) 26S proteasome non-ATPase regulatory subunit... 123 7e-29
RPN1_NEUCR (Q7S8R8) 26S proteasome regulatory subunit rpn-1 105 2e-23
RPN1_SCHPO (P87048) 26S proteasome regulatory subunit rpn1 (Prot... 92 3e-19
RPN1_YEAST (P38764) 26S proteasome regulatory subunit RPN1 (Prot... 81 4e-16
RPN1_CANGA (Q6FPV6) 26S proteasome regulatory subunit RPN1 75 4e-14
NRML_CAEEL (Q21434) NRAMP-like transporter smf-1 33 0.15
ML11_ARATH (Q9FI00) MLO-like protein 11 (AtMlo11) 32 0.34
NRM1_CEREL (P56436) Natural resistance-associated macrophage pro... 30 1.3
PODK_GIALA (P51776) Pyruvate, phosphate dikinase (EC 2.7.9.1) (P... 29 1.7
NRM1_CHICK (P51027) Natural resistance-associated macrophage pro... 29 1.7
MYS3_SCHPO (O14157) Myosin type II heavy chain 2 29 1.7
NRM1_SHEEP (P49280) Natural resistance-associated macrophage pro... 29 2.2
NRM1_BOVIN (Q27981) Natural resistance-associated macrophage pro... 28 3.8
NRM1_BISBI (Q95102) Natural resistance-associated macrophage pro... 28 3.8
VGLM_BUNGE (P12430) M polyprotein precursor [Contains: Glycoprot... 28 4.9
NRM1_BUBBU (Q27946) Natural resistance-associated macrophage pro... 28 4.9
ML14_ARATH (Q94KB1) MLO-like protein 14 (AtMlo14) 28 4.9
PCAQ_AGRT5 (P52668) HTH-type transcriptional regulator pcaQ (Pca... 27 6.5
METE_YERPE (Q8ZAL3) 5-methyltetrahydropteroyltriglutamate--homoc... 27 6.5
>PSD2_MOUSE (Q8VDM4) 26S proteasome non-ATPase regulatory subunit 2
(26S proteasome regulatory subunit RPN1) (26S proteasome
regulatory subunit S2) (26S proteasome subunit p97)
Length = 908
Score = 123 bits (309), Expect = 7e-29
Identities = 65/110 (59%), Positives = 83/110 (75%), Gaps = 11/110 (10%)
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDV 61
L++ ++ GK HYVLY LV AMQ P ML+T E L+PL V +RVGQAVDV
Sbjct: 804 LDVRNIILGKSHYVLYGLVAAMQ-----------PRMLVTFDEELRPLPVSVRVGQAVDV 852
Query: 62 VGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
VGQAG+PK ITGFQTH+T VLLA G+RAELATE+++P++P+LE FVIL++
Sbjct: 853 VGQAGKPKTITGFQTHTTPVLLAHGERAELATEEFLPVTPILEGFVILRK 902
>PSD2_HUMAN (Q13200) 26S proteasome non-ATPase regulatory subunit 2
(26S proteasome regulatory subunit RPN1) (26S proteasome
regulatory subunit S2) (26S proteasome subunit p97)
(Tumor necrosis factor type 1 receptor associated
protein 2) (55.11 protein)
Length = 908
Score = 123 bits (309), Expect = 7e-29
Identities = 65/110 (59%), Positives = 83/110 (75%), Gaps = 11/110 (10%)
Query: 2 LELPSV**GKYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDV 61
L++ ++ GK HYVLY LV AMQ P ML+T E L+PL V +RVGQAVDV
Sbjct: 804 LDVRNIILGKSHYVLYGLVAAMQ-----------PRMLVTFDEELRPLPVSVRVGQAVDV 852
Query: 62 VGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
VGQAG+PK ITGFQTH+T VLLA G+RAELATE+++P++P+LE FVIL++
Sbjct: 853 VGQAGKPKTITGFQTHTTPVLLAHGERAELATEEFLPVTPILEGFVILRK 902
>RPN1_NEUCR (Q7S8R8) 26S proteasome regulatory subunit rpn-1
Length = 883
Score = 105 bits (261), Expect = 2e-23
Identities = 57/101 (56%), Positives = 71/101 (69%), Gaps = 11/101 (10%)
Query: 11 KYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKK 70
K HY+LY++V AM P ML+T+ E+LKPL V +RVGQAVDVVGQAGRPK
Sbjct: 786 KSHYLLYWIVTAMH-----------PRMLVTLDEDLKPLTVNVRVGQAVDVVGQAGRPKT 834
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
ITG+QT ST VLL G+RAEL ++YI +S LE VIL++
Sbjct: 835 ITGWQTQSTPVLLGYGERAELEDDQYISLSSTLEGLVILRK 875
>RPN1_SCHPO (P87048) 26S proteasome regulatory subunit rpn1
(Proteasome non-ATPase subunit mts4) (19S regulatory cap
region of 26S protease subunit 2)
Length = 891
Score = 91.7 bits (226), Expect = 3e-19
Identities = 53/99 (53%), Positives = 69/99 (69%), Gaps = 12/99 (12%)
Query: 13 HYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKIT 72
H++LY + LA+ +P ML+T+GE+ + L V +RVGQAVDVVGQAGRPK IT
Sbjct: 796 HWLLYAITLAI-----------RPRMLITLGEDGQYLPVSVRVGQAVDVVGQAGRPKVIT 844
Query: 73 GFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
G+ TH+T VLL +RAELATE Y P++ LE VILK+
Sbjct: 845 GWVTHTTPVLLHHNERAELATEAYTPLTS-LEGIVILKK 882
>RPN1_YEAST (P38764) 26S proteasome regulatory subunit RPN1
(Proteasome non-ATPase subunit 1)
Length = 992
Score = 81.3 bits (199), Expect = 4e-16
Identities = 45/101 (44%), Positives = 63/101 (61%), Gaps = 11/101 (10%)
Query: 11 KYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKK 70
K+H + Y L + +P +L + + +P+ V +RVGQAV+ VGQAGRPKK
Sbjct: 895 KHHQLFYMLNAGI-----------RPKFILALNDEGEPIKVNVRVGQAVETVGQAGRPKK 943
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVILKE 111
ITG+ T ST VLL G+RAEL T++YI + +E VILK+
Sbjct: 944 ITGWITQSTPVLLNHGERAELETDEYISYTSHIEGVVILKK 984
>RPN1_CANGA (Q6FPV6) 26S proteasome regulatory subunit RPN1
Length = 983
Score = 74.7 bits (182), Expect = 4e-14
Identities = 37/77 (48%), Positives = 54/77 (70%)
Query: 35 KPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKITGFQTHSTSVLLAAGDRAELATE 94
+P ++T+ E + + V +R+GQAV+ VGQAG+PK ITG+ T ST VLL G+RAEL +
Sbjct: 899 RPKFIITLNEEGEQIKVNVRIGQAVETVGQAGKPKTITGWITQSTPVLLGHGERAELEND 958
Query: 95 KYIPISPVLEAFVILKE 111
+YI + +E VILK+
Sbjct: 959 EYISYTNNIEGVVILKK 975
>NRML_CAEEL (Q21434) NRAMP-like transporter smf-1
Length = 557
Score = 32.7 bits (73), Expect = 0.15
Identities = 27/105 (25%), Positives = 44/105 (41%), Gaps = 12/105 (11%)
Query: 11 KYHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVG 63
K + FL+L M ++ GY ++ KP +GE + ++VP G QA+ VVG
Sbjct: 192 KLELIFGFLILTMTVSFGYEFVVVKP----PIGEVISGMVVPWCAGCGKGEFMQAISVVG 247
Query: 64 QAGRPKKITGFQTHSTSVLLAAGDRAELA-TEKYIPISPVLEAFV 107
P + S + DR +A KY + + F+
Sbjct: 248 AVIMPHNLYLHSALVKSRRVDRKDRRRVAEANKYFTLESAIALFL 292
>ML11_ARATH (Q9FI00) MLO-like protein 11 (AtMlo11)
Length = 573
Score = 31.6 bits (70), Expect = 0.34
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 12/78 (15%)
Query: 11 KYHYVLYFLVLAMQLTLGY-------YILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVG 63
K HY++YF +L LG+ Y L ++ +G N K L+P R+ + + G
Sbjct: 402 KNHYLVYF-----RLLLGFAGQFLCSYSTLPLYALVTQMGTNYKAALIPQRIRETIRGWG 456
Query: 64 QAGRPKKITGFQTHSTSV 81
+A R K+ G ++V
Sbjct: 457 KATRRKRRHGLYGDDSTV 474
>NRM1_CEREL (P56436) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 548
Score = 29.6 bits (65), Expect = 1.3
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 14/99 (14%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P G L+ L +P G QAV +VG P
Sbjct: 198 FLITIMALTFGYEYVVARPAQ----GALLQGLFLPSCAGCGQPELLQAVGIVGAIIMPHN 253
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVIL 109
I HS+ V DR+ A + + ++EA + L
Sbjct: 254 I---YLHSSLVKSREVDRSRRADIREANMYFLIEATIAL 289
>PODK_GIALA (P51776) Pyruvate, phosphate dikinase (EC 2.7.9.1)
(Pyruvate, orthophosphate dikinase)
Length = 884
Score = 29.3 bits (64), Expect = 1.7
Identities = 19/56 (33%), Positives = 24/56 (41%), Gaps = 1/56 (1%)
Query: 41 TVGENLKPLLVPLRVGQAVDVVGQAGRPKKITGFQTHSTSVLLAAGDRAELATEKY 96
T G+N PLLV +R G AV + G + G S L A A A + Y
Sbjct: 86 TFGDNTNPLLVSVRSGAAVSMPGMMDTILNL-GLNDESVKGLAAVTGNARFAYDSY 140
>NRM1_CHICK (P51027) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 555
Score = 29.3 bits (64), Expect = 1.7
Identities = 26/80 (32%), Positives = 36/80 (44%), Gaps = 14/80 (17%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P E LK + +P G QAV +VG P
Sbjct: 206 FLITIMALTFGYEYVMVRPAQT----EVLKGIFLPYCPGCGREELLQAVGIVGAIIMPHN 261
Query: 71 ITGFQTHSTSVLLAAGDRAE 90
I HS+ V A DR++
Sbjct: 262 IF---LHSSLVKTRAIDRSK 278
>MYS3_SCHPO (O14157) Myosin type II heavy chain 2
Length = 2104
Score = 29.3 bits (64), Expect = 1.7
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query: 50 LVPLRVGQAVDVVGQAGRPKKITGFQT--HSTSVLLAAGDRAELATEKYIPISPVLEAF 106
L+ R+ Q++ V G++G K T + + TSV A+ ++ +K + +PVLEAF
Sbjct: 169 LLERRINQSILVTGESGAGKTETTKKVIQYLTSVTDASTSDSQQLEKKILETNPVLEAF 227
>NRM1_SHEEP (P49280) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 548
Score = 28.9 bits (63), Expect = 2.2
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 14/99 (14%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P G L+ L +P G QAV +VG P
Sbjct: 198 FLITIMALTFGYEYVVARPAQ----GALLQGLFLPSCPGCGQPELLQAVGIVGAIIMPHN 253
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVIL 109
I HS+ V DR+ A + + ++EA + L
Sbjct: 254 I---YLHSSLVKSREVDRSRRADIREANMYFLIEATIAL 289
>NRM1_BOVIN (Q27981) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 548
Score = 28.1 bits (61), Expect = 3.8
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 14/99 (14%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P G L+ L +P G QAV ++G P
Sbjct: 198 FLITIMALTFGYEYVVAQPAQ----GALLQGLFLPSCPGCGQPELLQAVGIIGAIIMPHN 253
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVIL 109
I HS+ V DR+ A + + ++EA + L
Sbjct: 254 I---YLHSSLVKSREVDRSRRADIREANMYFLIEATIAL 289
>NRM1_BISBI (Q95102) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 548
Score = 28.1 bits (61), Expect = 3.8
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 14/99 (14%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P G L+ L +P G QAV ++G P
Sbjct: 198 FLITIMALTFGYEYVVAQPAQ----GALLQGLFLPSCPGCGQPELLQAVGIIGAIIMPHN 253
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVIL 109
I HS+ V DR+ A + + ++EA + L
Sbjct: 254 I---YLHSSLVKSREVDRSRRADIREANMYFLIEATIAL 289
>VGLM_BUNGE (P12430) M polyprotein precursor [Contains: Glycoprotein
G2; Nonstructural protein NS-M; Glycoprotein G1]
Length = 1437
Score = 27.7 bits (60), Expect = 4.9
Identities = 25/97 (25%), Positives = 40/97 (40%), Gaps = 9/97 (9%)
Query: 13 HYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKIT 72
+Y+ + + L IL+Q TML+ GEN D VG P T
Sbjct: 452 YYIKIRNLKLIMLIFSIVILMQNATMLVVAGENCW-----TNTEIKADCVGPLIGPSACT 506
Query: 73 --GFQTHST--SVLLAAGDRAELATEKYIPISPVLEA 105
G +T+ T L+ A +L +KY+ + +E+
Sbjct: 507 NKGSKTYKTVAQELVTASKITQLDADKYVLLGDTIES 543
>NRM1_BUBBU (Q27946) Natural resistance-associated macrophage
protein 1 (NRAMP 1)
Length = 548
Score = 27.7 bits (60), Expect = 4.9
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 14/99 (14%)
Query: 18 FLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLVPLRVG-------QAVDVVGQAGRPKK 70
FL+ M LT GY ++ +P G L+ L +P G QAV ++G P
Sbjct: 198 FLITIMALTFGYEYVVAQPAQ----GALLQGLFLPSCRGCGQPELLQAVGIIGAIIMPHN 253
Query: 71 ITGFQTHSTSVLLAAGDRAELATEKYIPISPVLEAFVIL 109
I HS+ V DR+ A + + ++EA + L
Sbjct: 254 I---YLHSSLVKSREVDRSRRADIREANMYFLIEATIAL 289
>ML14_ARATH (Q94KB1) MLO-like protein 14 (AtMlo14)
Length = 554
Score = 27.7 bits (60), Expect = 4.9
Identities = 18/70 (25%), Positives = 35/70 (49%), Gaps = 7/70 (10%)
Query: 19 LVLAMQLTLGY-------YILLQKPTMLLTVGENLKPLLVPLRVGQAVDVVGQAGRPKKI 71
L++ ++L LG+ Y L ++ +G N K L+P RV + ++ G+A R K+
Sbjct: 400 LLVYLRLILGFSGQFLCSYSTLPLYALVTQMGTNYKAALLPQRVRETINGWGKATRRKRR 459
Query: 72 TGFQTHSTSV 81
G +++
Sbjct: 460 HGLYGDDSTI 469
>PCAQ_AGRT5 (P52668) HTH-type transcriptional regulator pcaQ (Pca
operon transcriptional activator)
Length = 311
Score = 27.3 bits (59), Expect = 6.5
Identities = 16/35 (45%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Query: 40 LTVGENLKPLLVPLRVGQAVDVVGQAGRPKKITGF 74
+ GEN LL LRVG VVG+ P+K+ GF
Sbjct: 129 IVTGENAV-LLEQLRVGDLDLVVGRLAAPQKMAGF 162
>METE_YERPE (Q8ZAL3)
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14) (Methionine synthase,
vitamin-B12 independent isozyme) (Cobalamin-independent
methionine synthase)
Length = 758
Score = 27.3 bits (59), Expect = 6.5
Identities = 13/40 (32%), Positives = 21/40 (52%)
Query: 12 YHYVLYFLVLAMQLTLGYYILLQKPTMLLTVGENLKPLLV 51
YHY++ Q LG+ LL + L +G +KP+L+
Sbjct: 123 YHYMVPEFQQGQQFKLGWTQLLDEVDEALALGHKIKPVLL 162
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.149 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,041,752
Number of Sequences: 164201
Number of extensions: 428652
Number of successful extensions: 1112
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1099
Number of HSP's gapped (non-prelim): 24
length of query: 112
length of database: 59,974,054
effective HSP length: 88
effective length of query: 24
effective length of database: 45,524,366
effective search space: 1092584784
effective search space used: 1092584784
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146719.10


