

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
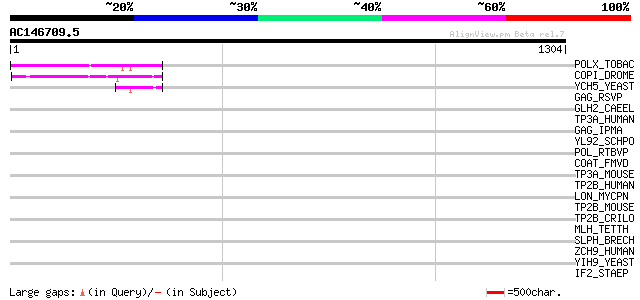
Query= AC146709.5 + phase: 0 /pseudo
(1304 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 209 4e-53
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 103 3e-21
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 49 1e-04
GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; C... 40 0.054
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 39 0.071
TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2) 39 0.093
GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains: ... 39 0.12
YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I 37 0.27
POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat pr... 37 0.35
COAT_FMVD (P09519) Probable coat protein 37 0.35
TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2) 36 0.60
TP2B_HUMAN (Q02880) DNA topoisomerase II, beta isozyme (EC 5.99.... 36 0.60
LON_MYCPN (P78025) ATP-dependent protease La (EC 3.4.21.53) 36 0.78
TP2B_MOUSE (Q64511) DNA topoisomerase II, beta isozyme (EC 5.99.... 35 1.0
TP2B_CRILO (Q64399) DNA topoisomerase II, beta isozyme (EC 5.99.... 35 1.3
MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC ... 35 1.7
SLPH_BRECH (P38538) Surface layer protein precursor (Hexagonal w... 34 2.3
ZCH9_HUMAN (Q8N567) Zinc finger CCHC domain containing protein 9 34 3.0
YIH9_YEAST (P40507) Hypothetical 41.6 kDa protein in SDS3-THS1 i... 33 3.9
IF2_STAEP (Q8CST4) Translation initiation factor IF-2 33 3.9
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 209 bits (532), Expect = 4e-53
Identities = 127/369 (34%), Positives = 203/369 (54%), Gaps = 19/369 (5%)
Query: 3 GSKWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDKAVSA 62
G K+++ KF G N F W+ +MR +LIQQ + L +++ + + +++++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQL 122
I L L D V+ + E TA +W +L+SLYM+K+L ++ LK+QLY M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 TEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRT 182
FN +I LAN+ V +E+EDK + LL +LP S++N T+L+GK TI L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 183 KELTKFKELKVDDSGEGL-NISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNP 241
E + K ++ G+ L RGRS+ + S ++ +SK C+ C+ P
Sbjct: 182 NEKMR---KKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQP 238
Query: 242 GHFKKDCPERKDNGGGESSVQ-----IASKDEGYESAGALTVTSWE-------PEKSWVL 289
GHFK+DCP + G GE+S Q A+ + ++ L + E PE WV+
Sbjct: 239 GHFKRDCPNPR-KGKGETSGQKNDDNTAAMVQNNDNV-VLFINEEEECMHLSGPESEWVV 296
Query: 290 DSGCSYHICPRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKNVRYI 349
D+ S+H P ++ F + G V++GN KI G+G I +K +LK+VR++
Sbjct: 297 DTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDVRHV 356
Query: 350 PELKRNLIS 358
P+L+ NLIS
Sbjct: 357 PDLRMNLIS 365
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 103 bits (257), Expect = 3e-21
Identities = 98/370 (26%), Positives = 169/370 (45%), Gaps = 26/370 (7%)
Query: 4 SKWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMSAHLTPAEKTEMNDKAV--- 60
+K +I+ F G + +WK ++RA+L +Q ++ + G L P E + KA
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDG-------LMPNEVDDSWKKAERCA 55
Query: 61 -SAIILCLGDKVLREVSREATAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIM 119
S II L D L + + TA + LD++Y KSLA + L+++L ++ ++
Sbjct: 56 KSTIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLL 115
Query: 120 EQLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEEVQAA 179
F+++I +L +E+ DK+ HLL LP ++ + E +TL V+
Sbjct: 116 SHFHIFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNR 175
Query: 180 LRTKELTKFKELKVDDSGEGLNISRGRSHNKGKG---KGKNSRSKSRSKGDGNKTQYKCF 236
L +E+ K K D S + +N ++N K K + ++ K KG+ +K + KC
Sbjct: 176 LLDQEI-KIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGN-SKYKVKCH 233
Query: 237 ICHNPGHFKKDCPERK-----DNGGGESSVQIASKDEGYESAGALTVTSWEPEKSWVLDS 291
C GH KKDC K N E VQ A+ + TS +VLDS
Sbjct: 234 HCGREGHIKKDCFHYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDS 293
Query: 292 GCSYHICPRKE-YFETLELKEGGVVRLG-NNKACKIQGMGTIRLKMFDDRDFLLKNVRYI 349
G S H+ + Y +++E+ + + + G +RL+ +D + L++V +
Sbjct: 294 GASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLR--NDHEITLEDVLFC 351
Query: 350 PELKRNLISI 359
E NL+S+
Sbjct: 352 KEAAGNLMSV 361
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 48.5 bits (114), Expect = 1e-04
Identities = 31/121 (25%), Positives = 50/121 (40%), Gaps = 15/121 (12%)
Query: 249 PERKDNGGGESSVQIASKDEGYESAGALTVTSW----------EPEKSWVLDSGCSYHIC 298
P K + +SV I + ++AG +T SW W+ D+GC+ H+C
Sbjct: 31 PNDKTSRSSSASVAIPDYETQGQTAGQITPKSWLCMLSSTVPATKSSEWIFDTGCTSHMC 90
Query: 299 PRKEYFETLELKEGGVVRLGNNKACKIQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLIS 358
+ F + G + I G GT+ + L +V Y+P+L NLIS
Sbjct: 91 HDRSIFSSFTRSSRKDFVRGVGGSIPIMGSGTVNIGTVQ-----LHDVSYVPDLPVNLIS 145
Query: 359 I 359
+
Sbjct: 146 V 146
>GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; Core
protein p2A; Core protein p2B; Core protein p10; Capsid
protein p27; Inner coat protein p12; Protease p15 (EC
3.4.23.-)]
Length = 701
Score = 39.7 bits (91), Expect = 0.054
Identities = 12/48 (25%), Positives = 26/48 (54%)
Query: 218 SRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIAS 265
+R + G G + + C+ C +PGH++ CP+++ +G Q+ +
Sbjct: 492 NRERDGQTGSGGRARGLCYTCGSPGHYQAQCPKKRKSGNSRERCQLCN 539
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 39.3 bits (90), Expect = 0.071
Identities = 21/58 (36%), Positives = 23/58 (39%), Gaps = 2/58 (3%)
Query: 197 GEGLNISRGRSHNKGKGKGKNSRSKSRSKG--DGNKTQYKCFICHNPGHFKKDCPERK 252
G+ G S G G G NS G D + CF C PGH DCPE K
Sbjct: 219 GKSGGFGGGNSGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPK 276
Score = 36.2 bits (82), Expect = 0.60
Identities = 20/58 (34%), Positives = 24/58 (40%), Gaps = 9/58 (15%)
Query: 195 DSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 252
+SG G G S+ G G G R + + CF C PGH DCPE K
Sbjct: 342 ESGFGSGGFGGNSNGFGSGGGGQDRGERNNN---------CFNCQQPGHRSNDCPEPK 390
>TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1001
Score = 38.9 bits (89), Expect = 0.093
Identities = 17/49 (34%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Query: 204 RGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 252
RGR+ + + + K R+ S G K KC +CH PGH + CP+ +
Sbjct: 954 RGRTL-ESEARSKRPRASSSDMGSTAKKPRKCSLCHQPGHTRPFCPQNR 1001
>GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains:
Protease (EC 3.4.23.-)]
Length = 827
Score = 38.5 bits (88), Expect = 0.12
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 8/57 (14%)
Query: 218 SRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASK-DEGYESA 273
S+++S S+ D Q CF C PGHFKKDC GG ++ + SK +GY A
Sbjct: 447 SQNRSMSRND----QRTCFNCGKPGHFKKDCRAPDKQGG---TLTLCSKCGKGYHRA 496
>YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I
Length = 218
Score = 37.4 bits (85), Expect = 0.27
Identities = 24/80 (30%), Positives = 38/80 (47%), Gaps = 10/80 (12%)
Query: 196 SGEGLNISRGRS--HNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKD 253
S + + S G S +++ + K ++ + R N+ ++ CF C GH +DCPE KD
Sbjct: 39 SQDNMKASFGSSKRYDERQKKKRSEYRRLRRINQRNRDKF-CFACRQQGHIVQDCPEAKD 97
Query: 254 N-------GGGESSVQIASK 266
N G E S+ SK
Sbjct: 98 NVSICFRCGSKEHSLNACSK 117
>POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat
protein; Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1675
Score = 37.0 bits (84), Expect = 0.35
Identities = 47/189 (24%), Positives = 73/189 (37%), Gaps = 36/189 (19%)
Query: 218 SRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDN--------GGGESSVQIASKDEG 269
+R K+ + K + +C+IC + H CP R N G E V IAS DE
Sbjct: 757 NRRKNYVRRPSIKKKCRCYICQDENHLANRCPRRYTNQARASLIDGLDEDIVSIASDDED 816
Query: 270 YES-------AGALTVTSWEPEKSWVLDSGCSYHICPRKEYFETLELKEGGVVRLGNNKA 322
E+ + +S E E +W + G +C YF NK
Sbjct: 817 IENFLEIIELDEFIAHSSQEHEHTWEI-GGKKDKVCEICSYFTDY------------NKT 863
Query: 323 CKIQGMGTIRLKMFDDRDFLLKNVRYIPELKRNLISISMFDGLELNVV**EF---LMVHW 379
+ T K D+ L + E+K+ +M D L+LNV EF ++ H
Sbjct: 864 VSCKTCETQYCKTCSDQLAL-----EVTEVKKPTKEETMIDDLKLNVKNLEFRVTILEHK 918
Query: 380 LWLRGLKYK 388
+ ++ L+ K
Sbjct: 919 VEMQNLQDK 927
>COAT_FMVD (P09519) Probable coat protein
Length = 489
Score = 37.0 bits (84), Expect = 0.35
Identities = 24/88 (27%), Positives = 41/88 (46%), Gaps = 4/88 (4%)
Query: 183 KELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 242
K+ +K+K K + + L + R GK K K +G + + +C+IC G
Sbjct: 362 KKSSKYKAYKKKKTLKKLWKKKKRKFTPGKYFSKKKPEKFCPQG---RKKCRCWICTEEG 418
Query: 243 HFKKDCPERKDNGGGESSVQIASKDEGY 270
H+ +CP RK + + + I +EGY
Sbjct: 419 HYANECPNRKSH-QEKVKILIHGMNEGY 445
>TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1003
Score = 36.2 bits (82), Expect = 0.60
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 204 RGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 252
RG++ + + K R+ S G K KC +CH PGH + CP+ +
Sbjct: 956 RGKAQ-RPEAASKRPRAGSSDAGSTVKKPRKCSLCHQPGHTRTFCPQNR 1003
>TP2B_HUMAN (Q02880) DNA topoisomerase II, beta isozyme (EC 5.99.1.3)
Length = 1626
Score = 36.2 bits (82), Expect = 0.60
Identities = 30/113 (26%), Positives = 45/113 (39%), Gaps = 10/113 (8%)
Query: 130 DDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFK 189
DD A D N ED V L ++ + + K+G + + V R + K
Sbjct: 1459 DDSAKFDSNEEDSASVFSPSFGLKQT-DKVPSKTVAAKKGKPSSDTVPKPKRAPKQKKVV 1517
Query: 190 ELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 242
E DS I + + KGKG+G R S S+ +G+ +NPG
Sbjct: 1518 EAVNSDSDSEFGIPKKTTTPKGKGRGAKKRKASGSENEGD---------YNPG 1561
>LON_MYCPN (P78025) ATP-dependent protease La (EC 3.4.21.53)
Length = 795
Score = 35.8 bits (81), Expect = 0.78
Identities = 29/112 (25%), Positives = 56/112 (49%), Gaps = 5/112 (4%)
Query: 64 ILCLGDKVLREVSREATAV---SMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIME 120
I+ G + L +S EA A S ++K+D+ K + Q +QQ FY + + I E
Sbjct: 205 IIGSGIEDLGHISEEARAKQRESEFDKIDNRITRK--VNEQLSRQQRDFYLREKLRVIRE 262
Query: 121 QLTEFNKIIDDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTIT 172
++ +K D+++NI LE+ H+ + ++F+++ +E T+T
Sbjct: 263 EIGMTSKKEDEVSNIRKKLEENPYPEHIKKRILSELDHFENSSSSSQESTLT 314
>TP2B_MOUSE (Q64511) DNA topoisomerase II, beta isozyme (EC 5.99.1.3)
Length = 1612
Score = 35.4 bits (80), Expect = 1.0
Identities = 30/113 (26%), Positives = 44/113 (38%), Gaps = 10/113 (8%)
Query: 130 DDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFK 189
DD A D N ED V L ++ + + K+G + A R + K
Sbjct: 1446 DDSAKFDSNEEDTASVFAPSFGLKQT-DKLPSKTVAAKKGKPPSDTAPKAKRAPKQKKIV 1504
Query: 190 ELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 242
E DS I + + KGKG+G R S S+ +G+ +NPG
Sbjct: 1505 ETINSDSDSEFGIPKKTTTPKGKGRGAKKRKASGSENEGD---------YNPG 1548
>TP2B_CRILO (Q64399) DNA topoisomerase II, beta isozyme (EC 5.99.1.3)
Length = 1612
Score = 35.0 bits (79), Expect = 1.3
Identities = 30/113 (26%), Positives = 44/113 (38%), Gaps = 10/113 (8%)
Query: 130 DDLANIDVNLEDEDKVLHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFK 189
DD A D N ED V L ++ + + K+G + A R + K
Sbjct: 1446 DDSAKFDSNEEDTASVFTPSFGLKQT-DKVPSKTVAAKKGKPPSDTAPKAKRAPKQKKVV 1504
Query: 190 ELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 242
E DS I + + KGKG+G R S S+ +G+ +NPG
Sbjct: 1505 ETVNSDSDSEFGIPKKTTTPKGKGRGAKKRKASGSENEGD---------YNPG 1548
>MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC LH)
[Contains: Micronuclear linker histone-alpha;
Micronuclear linker histone-beta; Micronuclear linker
histone-delta; Micronuclear linker histone-gamma]
Length = 633
Score = 34.7 bits (78), Expect = 1.7
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 7/56 (12%)
Query: 181 RTKELTKFKELKVDDSGEGLNISRGRSHNKGKGK----GKNSRSKSRSKGDGNKTQ 232
RTK+ K SG S+G+S +K KGK GKNS+S+S SK N Q
Sbjct: 400 RTKKANNKSASKASKSGSK---SKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQ 452
Score = 32.7 bits (73), Expect = 6.6
Identities = 26/107 (24%), Positives = 48/107 (44%), Gaps = 4/107 (3%)
Query: 186 TKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRSKGDG-NKTQYKCFICHNPGHF 244
+K K S + + S+ + N G+ SRS+S+SK + NK K + P
Sbjct: 508 SKSKSKSASKSRKNASKSKKDTTNHGRQTRSKSRSESKSKSEAPNKPSNKMEVIEQP--- 564
Query: 245 KKDCPERKDNGGGESSVQIASKDEGYESAGALTVTSWEPEKSWVLDS 291
K++ +RK S + S + + + +T+ +P+K+ DS
Sbjct: 565 KEESSDRKRRESRSQSAKKTSDKKSKNRSDSKKMTAEDPKKNNAEDS 611
>SLPH_BRECH (P38538) Surface layer protein precursor (Hexagonal wall
protein) (HWP)
Length = 1116
Score = 34.3 bits (77), Expect = 2.3
Identities = 33/140 (23%), Positives = 56/140 (39%), Gaps = 10/140 (7%)
Query: 110 YRMVESKPIMEQLTEFNKIIDDLANI---------DVNLEDEDKVLHLLCALPRSFENFK 160
YR+ E I T FN +D L+ I V L D ++V +L ++ +
Sbjct: 381 YRLTEDTKITYNFTRFNDPVDALSKIYKDNDTFGVKVVLNDNNEVAYLHIIDDQTIDKSV 440
Query: 161 DTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRS 220
+ YG + ++ + + + +KF +L+ D G+ + K G K S
Sbjct: 441 KGVKYGSKVISKIDADKKKITNLDNSKFSDLEDQDEGKDFLVFLDGQPAK-LGDLKESDV 499
Query: 221 KSRSKGDGNKTQYKCFICHN 240
S DG+K +Y F N
Sbjct: 500 YSVYYADGDKDKYLVFANRN 519
>ZCH9_HUMAN (Q8N567) Zinc finger CCHC domain containing protein 9
Length = 271
Score = 33.9 bits (76), Expect = 3.0
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 12/81 (14%)
Query: 213 GKGKNSRSKSRSKGDGNKTQY---KCFICHNPGHFKKDCPER-----KDNGGGE--SSVQ 262
G ++ +K ++K D ++ KCF+C GH + CP+ D GG + SV+
Sbjct: 161 GSTEHEITKCKAKVDPALGEFPFAKCFVCGEMGHLSRSCPDNPKGLYADGGGCKLCGSVE 220
Query: 263 IASKD--EGYESAGALTVTSW 281
KD E S +TV W
Sbjct: 221 HLKKDCPESQNSERMVTVGRW 241
>YIH9_YEAST (P40507) Hypothetical 41.6 kDa protein in SDS3-THS1
intergenic region
Length = 360
Score = 33.5 bits (75), Expect = 3.9
Identities = 13/36 (36%), Positives = 21/36 (58%)
Query: 219 RSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKDN 254
++K ++GD + C+ C N GHF DC ER+ +
Sbjct: 159 KTKDANQGDFDFQTVFCYNCGNAGHFGDDCAERRSS 194
>IF2_STAEP (Q8CST4) Translation initiation factor IF-2
Length = 720
Score = 33.5 bits (75), Expect = 3.9
Identities = 43/172 (25%), Positives = 72/172 (41%), Gaps = 16/172 (9%)
Query: 113 VESKPIMEQLTEFN----KIIDDLANIDVNLEDEDKVLH--LLCALPRSFE--NFKDTML 164
+ K I E E N +IID+L +++V + + + L + AL + F+ KDT
Sbjct: 1 MSKKRIYEYAKELNLKSKEIIDELKSMNVEVSNHMQALEEEQIKALDKKFKASQAKDT-- 58
Query: 165 YGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNISRGRSHNKGKGKGKNSRSKSRS 224
K+ T + K+ + KE K + +NKGK + KN+++
Sbjct: 59 -NKQNT---QNNHQKSNNKQNSNDKE-KQQSKNNSKPTKKKEQNNKGKQQNKNNKTNKNQ 113
Query: 225 KGDGNKTQYKCFICHNPGHFKKDCPERKDNGGGESSVQIASKDEGYESAGAL 276
K + NK K N K+ P + G + ++A K ESAG +
Sbjct: 114 KNNKNKKNNKNNKPQNEVEETKEMPSKITYQEGITVGELAEK-LNVESAGII 164
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.357 0.158 0.567
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 124,496,088
Number of Sequences: 164201
Number of extensions: 4504552
Number of successful extensions: 21463
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 21389
Number of HSP's gapped (non-prelim): 72
length of query: 1304
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1182
effective length of database: 39,941,532
effective search space: 47210890824
effective search space used: 47210890824
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146709.5


