

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146706.7 + phase: 0 /pseudo
(617 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
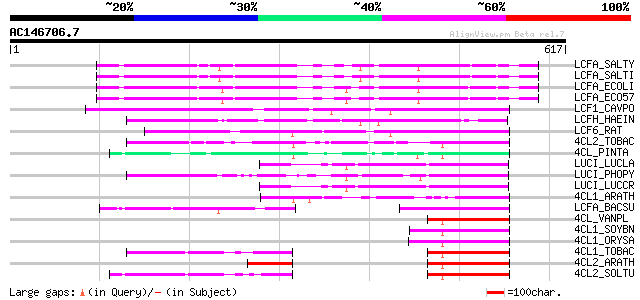
Score E
Sequences producing significant alignments: (bits) Value
LCFA_SALTY (P63521) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 134 5e-31
LCFA_SALTI (P63522) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 134 5e-31
LCFA_ECOLI (P29212) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 134 7e-31
LCFA_ECO57 (Q8XDR6) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 133 1e-30
LCF1_CAVPO (Q9JID6) Long-chain-fatty-acid--CoA ligase 1 (EC 6.2.... 133 1e-30
LCFH_HAEIN (P44446) Putative long-chain-fatty-acid--CoA ligase (... 127 7e-29
LCF6_RAT (P33124) Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.... 105 5e-22
4CL2_TOBAC (O24146) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 104 8e-22
4CL_PINTA (P41636) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL) (... 101 7e-21
LUCI_LUCLA (Q01158) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 97 2e-19
LUCI_PHOPY (P08659) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 96 2e-19
LUCI_LUCCR (P13129) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 95 5e-19
4CL1_ARATH (Q42524) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 88 8e-17
LCFA_BACSU (P94547) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 87 1e-16
4CL_VANPL (O24540) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL) (... 87 1e-16
4CL1_SOYBN (P31686) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 85 6e-16
4CL1_ORYSA (P17814) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 85 6e-16
4CL1_TOBAC (O24145) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 84 1e-15
4CL2_ARATH (Q9S725) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 84 1e-15
4CL2_SOLTU (P31685) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 82 5e-15
>LCFA_SALTY (P63521) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 561
Score = 134 bits (338), Expect = 5e-31
Identities = 139/508 (27%), Positives = 231/508 (45%), Gaps = 78/508 (15%)
Query: 97 DEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRV-IGVSPNE 155
D ++++ +++ +A +Y D+ A ++ MT+++LE+ FA L+ +G+ +
Sbjct: 20 DRYQSLVELFEHAATRYADQPAFVNM----GEVMTFRKLEERSRAFAAYLQQGLGLKKGD 75
Query: 156 KIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNR 215
++AL N ++ VA G++ G I V + EL N S + A+ + + +
Sbjct: 76 RVALMMPNLLQYPVALFGILRAGMIVVNVNPLYTPRELEHQLNDSGAAAIIIVS-NFAHT 134
Query: 216 IAKAFDLKASMRFVIL------LWGEKSCLVNEGSKEVPIFTFTEIMHLGRG-SRRLFES 268
+ K + K S++ VIL L K +VN K + HL S R
Sbjct: 135 LEKVVE-KTSVQHVILTRMGDQLSTAKGTVVNFVVKYIK--RLVPKYHLPDAISFRSALQ 191
Query: 269 HDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*V 328
H R YV + ++D+A L YT GTTG KG MLTH+N+L + + A+
Sbjct: 192 HGYRMQYVKPEVVAEDLAFLQYTGGTTGVAKGAMLTHRNMLANLEQVKATYG-------- 243
Query: 329 FHFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYE 388
P ++ ++L V T+ + + ALT M C E
Sbjct: 244 --------PLLHPGKELVV-------------TALPLYHIFALT--------MNCLLFIE 274
Query: 389 ----GKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGI 444
+T + P + + A+ T + ++ L L+ +K + FS +
Sbjct: 275 LGGQNLLITNPRDIPGLVKEL-----AKYPFTAMTGVNTLFNALLNNKEFQQLDFSSLHL 329
Query: 445 SGGGSLPSH---VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNV-IGSVGHPLKHTEF 500
S GG +P +R+ + G L GYGLTE +P+++ + GS+G P+ TE
Sbjct: 330 SAGGGMPVQNVVAERWVKLTGQYLLEGYGLTECAPLVSVNPHDIDYHSGSIGLPVPSTEA 389
Query: 501 KVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHS 560
K+VD + EV P G G L V+GPQ+M GY++ P AT++ I KDGWL+TGDI +
Sbjct: 390 KLVDDDDNEVAP-GEAGELCVKGPQVMLGYWQRPDATDEII-KDGWLHTGDIAVM----- 442
Query: 561 SGRSRNCGGVIVVEGRAKDTIVLSSEKV 588
+ G + + R KD I++S V
Sbjct: 443 -----DEDGFLRIVDRKKDMILVSGFNV 465
>LCFA_SALTI (P63522) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 561
Score = 134 bits (338), Expect = 5e-31
Identities = 139/508 (27%), Positives = 231/508 (45%), Gaps = 78/508 (15%)
Query: 97 DEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRV-IGVSPNE 155
D ++++ +++ +A +Y D+ A ++ MT+++LE+ FA L+ +G+ +
Sbjct: 20 DRYQSLVELFEHAATRYADQPAFVNM----GEVMTFRKLEERSRAFAAYLQQGLGLKKGD 75
Query: 156 KIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNR 215
++AL N ++ VA G++ G I V + EL N S + A+ + + +
Sbjct: 76 RVALMMPNLLQYPVALFGILRAGMIVVNVNPLYTPRELEHQLNDSGAAAIIIVS-NFAHT 134
Query: 216 IAKAFDLKASMRFVIL------LWGEKSCLVNEGSKEVPIFTFTEIMHLGRG-SRRLFES 268
+ K + K S++ VIL L K +VN K + HL S R
Sbjct: 135 LEKVVE-KTSVQHVILTRMGDQLSTAKGTVVNFVVKYIK--RLVPKYHLPDAISFRSALQ 191
Query: 269 HDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*V 328
H R YV + ++D+A L YT GTTG KG MLTH+N+L + + A+
Sbjct: 192 HGYRMQYVKPEVVAEDLAFLQYTGGTTGVAKGAMLTHRNMLANLEQVKATYG-------- 243
Query: 329 FHFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYE 388
P ++ ++L V T+ + + ALT M C E
Sbjct: 244 --------PLLHPGKELVV-------------TALPLYHIFALT--------MNCLLFIE 274
Query: 389 ----GKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGI 444
+T + P + + A+ T + ++ L L+ +K + FS +
Sbjct: 275 LGGQNLLITNPRDIPGLVKEL-----AKYPFTAMTGVNTLFNALLNNKEFQQLDFSSLHL 329
Query: 445 SGGGSLPSH---VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNV-IGSVGHPLKHTEF 500
S GG +P +R+ + G L GYGLTE +P+++ + GS+G P+ TE
Sbjct: 330 SAGGGMPVQNVVAERWVKLTGQYLLEGYGLTECAPLVSVNPHDIDYHSGSIGLPVPSTEA 389
Query: 501 KVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHS 560
K+VD + EV P G G L V+GPQ+M GY++ P AT++ I KDGWL+TGDI +
Sbjct: 390 KLVDDDDNEVAP-GEAGELCVKGPQVMLGYWQRPDATDEII-KDGWLHTGDIAVM----- 442
Query: 561 SGRSRNCGGVIVVEGRAKDTIVLSSEKV 588
+ G + + R KD I++S V
Sbjct: 443 -----DEDGFLRIVDRKKDMILVSGFNV 465
>LCFA_ECOLI (P29212) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 561
Score = 134 bits (337), Expect = 7e-31
Identities = 134/506 (26%), Positives = 228/506 (44%), Gaps = 74/506 (14%)
Query: 97 DEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRV-IGVSPNE 155
D ++++ D++ S +Y D+ A ++ MT+++LE+ FA L+ +G+ +
Sbjct: 20 DRYQSLVDMFEQSVARYADQPAFVNM----GEVMTFRKLEERSRAFAAYLQQGLGLKKGD 75
Query: 156 KIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNR 215
++AL N ++ VA G++ G I V + EL N S + A+ + + +
Sbjct: 76 RVALMMPNLLQYPVALFGILRAGMIVVNVNPLYTPRELEHQLNDSGASAIVIVS-NFAHT 134
Query: 216 IAKAFDLKASMRFVILLWGE-----KSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHD 270
+ K D A ++ G+ K +VN K + HL H+
Sbjct: 135 LEKVVDKTAVQHVILTRMGDQLSTAKGTVVNFVVKYIK--RLVPKYHLPDAISFRSALHN 192
Query: 271 A-RKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*VF 329
R YV + +D+A L YT GTTG KG MLTH+N+L + + A+
Sbjct: 193 GYRMQYVKPELVPEDLAFLQYTGGTTGVAKGAMLTHRNMLANLEQVNATYG--------- 243
Query: 330 HFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTF---IRVSLGYMECKRI 386
P ++ ++L V T+ + + ALT + + LG
Sbjct: 244 -------PLLHPGKELVV-------------TALPLYHIFALTINCLLFIELGG------ 277
Query: 387 YEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISG 446
+ +T + P + + A+ T + ++ L L+ +K + FS +S
Sbjct: 278 -QNLLITNPRDIPGLVKEL-----AKYPFTAITGVNTLFNALLNNKEFQQLDFSSLHLSA 331
Query: 447 GGSLPSH---VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNV-IGSVGHPLKHTEFKV 502
GG +P +R+ + G L GYGLTE +P+++ + GS+G P+ TE K+
Sbjct: 332 GGGMPVQQVVAERWVKLTGQYLLEGYGLTECAPLVSVNPYDIDYHSGSIGLPVPSTEAKL 391
Query: 503 VDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSG 562
VD + EV PPG G L V+GPQ+M GY++ P AT++ I K+GWL+TGDI +
Sbjct: 392 VDDDDNEV-PPGQPGELCVKGPQVMLGYWQRPDATDEII-KNGWLHTGDIAVM------- 442
Query: 563 RSRNCGGVIVVEGRAKDTIVLSSEKV 588
+ G + + R KD I++S V
Sbjct: 443 ---DEEGFLRIVDRKKDMILVSGFNV 465
>LCFA_ECO57 (Q8XDR6) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 561
Score = 133 bits (335), Expect = 1e-30
Identities = 135/506 (26%), Positives = 227/506 (44%), Gaps = 74/506 (14%)
Query: 97 DEWKTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRV-IGVSPNE 155
D ++++ D++ S +Y D+ A ++ MT+++LE+ FA L+ +G+ +
Sbjct: 20 DRYQSLVDMFEQSVARYADQPAFVNM----GEVMTFRKLEERSRAFAAYLQQGLGLKKGD 75
Query: 156 KIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNR 215
++AL N ++ VA G++ G I V + EL N S + A+ + + +
Sbjct: 76 RVALMMPNLLQYPVALFGILRAGMIVVNVNPLYTPRELEHQLNDSGASAIVIVS-NFAHT 134
Query: 216 IAKAFDLKASMRFVILLWGE-----KSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHD 270
+ K D A ++ G+ K LVN K + HL H+
Sbjct: 135 LEKVVDKTAVQHVILTRMGDQLSTAKGTLVNFVVKYIK--RLVPKYHLPDAISFRSALHN 192
Query: 271 A-RKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*VF 329
R YV + +D+A L YT GTTG KG MLTH+N+L + + A+
Sbjct: 193 GYRMQYVKPELVPEDLAFLQYTGGTTGVAKGAMLTHRNMLANLEQVNATYG--------- 243
Query: 330 HFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTF---IRVSLGYMECKRI 386
P ++ ++L V T+ + + ALT + + LG
Sbjct: 244 -------PLLHPGKELVV-------------TALPLYHIFALTINCLLFIELGG------ 277
Query: 387 YEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISG 446
+ +T + P + + A+ T + ++ L L+ +K + FS +S
Sbjct: 278 -QNLLITNPRDIPGLVKEL-----AKYPFTAITGVNTLFNALLNNKEFQQLDFSSLHLSA 331
Query: 447 GGSLPSH---VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNV-IGSVGHPLKHTEFKV 502
GG +P +R+ + G L GYGLTE +P+++ + GS+G P+ TE K+
Sbjct: 332 GGGMPVQQVVAERWVKLTGQYLLEGYGLTECAPLVSVNPYDIDYHSGSIGLPVPSTEAKL 391
Query: 503 VDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWIAAYHSSG 562
VD + EV PG G L VRGPQ+M GY++ P AT++ I K+GWL+TGDI +
Sbjct: 392 VDDDDNEV-SPGQPGELCVRGPQVMLGYWQRPDATDEII-KNGWLHTGDIAVM------- 442
Query: 563 RSRNCGGVIVVEGRAKDTIVLSSEKV 588
+ G + + R KD I++S V
Sbjct: 443 ---DEEGFLRIVDRKKDMILVSGFNV 465
>LCF1_CAVPO (Q9JID6) Long-chain-fatty-acid--CoA ligase 1 (EC
6.2.1.3) (Long-chain acyl-CoA synthetase 1) (LACS 1)
(Palmitoyl-CoA ligase)
Length = 698
Score = 133 bits (335), Expect = 1e-30
Identities = 131/490 (26%), Positives = 225/490 (45%), Gaps = 36/490 (7%)
Query: 85 SSLLSGNDGVVS--DEWKTVPDIWRSSAEKYGDKIALIDPYHDPP-STMTYKQLEDAILD 141
S+LL ++ +V D+ +T+ D+++ + + L D P ++YKQ+ED
Sbjct: 73 STLLDSDEPLVYFYDDVRTLYDVFQRGIQVSNNGPCLGSRKPDQPYEWLSYKQVEDLSEC 132
Query: 142 FAEGLRVIG--VSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNH 199
L G SP++ I +FA N W++ +Q A + V + + I N
Sbjct: 133 IGSALLQKGFQASPDQFIGIFAQNRPEWVIIEQACFAYSMVVVPLYDTLGADAITYIVNK 192
Query: 200 SESIALAVDNPEMFNRIAKAFDLKAS--MRFVILLWGEKSCLVNEGSK-EVPIFTFTEIM 256
+E + D PE + ++ + K + ++ ++++ S LV +G K V + + +
Sbjct: 193 AELSVIFADKPEKARILLESVENKLTPGLKIIVVMDSYGSELVEQGKKCGVEVISLKAME 252
Query: 257 HLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKY- 315
LGR +RR + + DD+A + +TSGTTGNPKG M+TH+N++ S +
Sbjct: 253 GLGRANRRKPKPPEP-----------DDLAVICFTSGTTGNPKGAMITHKNVVSDCSAFV 301
Query: 316 -AASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQ---RQISTSSLVRKLVAL 371
A A + + + F+ P + E+L L G + Q L+ L AL
Sbjct: 302 KATEKALVLNASDIHISFL---PLAHMYEQLLQCVMLCHGAKIGFFQGDIRLLMDDLKAL 358
Query: 372 TFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPI---HMLAKKL 428
+ R+++ N +L LD+ R A + I + + KL
Sbjct: 359 QPTIFPVVPRLLNRMFDRIFAQANTTVKRWL---LDFASKRKEAELRSGIIRNNSVWDKL 415
Query: 429 VYSKIHSAIGFS-KAGISGGGSLPSHVDRFFE-AIGVTLQNGYGLTETSPVIAARRLSCN 486
++ KI S++G + ++G + + V F A+G GYG TE + +
Sbjct: 416 IFHKIQSSLGGKVRLMVTGAAPVSATVLTFLRAALGCQFYEGYGQTECTAGCSLSVPGDW 475
Query: 487 VIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGW 546
G VG P+ K+VD E + +G + V+GP + KGY K+P+ T +A+DKDGW
Sbjct: 476 TAGHVGAPMPCNFIKLVDVEEMNYMAAMGEGEVCVKGPNVFKGYLKDPAKTAEALDKDGW 535
Query: 547 LNTGDIG-WI 555
L+TGDIG W+
Sbjct: 536 LHTGDIGKWL 545
>LCFH_HAEIN (P44446) Putative long-chain-fatty-acid--CoA ligase (EC
6.2.1.3) (Long-chain acyl-CoA synthetase) (LACS)
Length = 607
Score = 127 bits (320), Expect = 7e-29
Identities = 120/441 (27%), Positives = 197/441 (44%), Gaps = 47/441 (10%)
Query: 130 MTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSS 189
+++K ++ + + L + +KIA+FA N RW + D + AI V + ++
Sbjct: 43 ISWKNFQEQLNQLSRALLAHNIDVQDKIAIFAHNMERWTIVDIATLQIRAITVPIYATNT 102
Query: 190 IEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPI 249
++ I NH++ L V + E +++ + ++ ++ + KS + + ++
Sbjct: 103 AQQAEFILNHADVKILFVGDQEQYDQTLEIAHHCPKLQKIVAM---KSTI--QLQQDPLS 157
Query: 250 FTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLL 309
T+ + G +++ + + + D+ T++YTSGTTG PKGVML + NL
Sbjct: 158 CTWESFIKTGSNAQQDELTQRLNQKQL------SDLFTIIYTSGTTGEPKGVMLDYANLA 211
Query: 310 HQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSG-IQRQISTSSLVRKL 368
HQ+ + SL + + + P + E+ L+ G I + ++ VR
Sbjct: 212 HQLETHDLSLNVTDQDISLSFL-----PFSHIFERAWAAYILHRGAILCYLEDTNQVRS- 265
Query: 369 VALTFIRVSLGYMECKRIYE---GKCLTKNQKSPSYLYAMLDW------------LGARI 413
ALT IR +L R YE L K QK+P M W +
Sbjct: 266 -ALTEIRPTL-MCAVPRFYEKIYAAVLDKVQKAPKLRQIMFHWAISVGQKYFDLRANNKA 323
Query: 414 IATILFPIHMLAKKLVYSKIHSAIGFS-KAGISGGGSLPSHVDRFFEAIGVTLQNGYGLT 472
I +L LA KLV SK+ +G K GG L + FF AIG+ ++ GYG+T
Sbjct: 324 IPFLLKKQFALADKLVLSKLRQLLGGRIKMMPCGGAKLEPAIGLFFHAIGINIKLGYGMT 383
Query: 473 ETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYK 532
ET+ ++ S+G + E K+ E E+L VRG +MKGYYK
Sbjct: 384 ETTATVSCWHDFQFNPNSIGTLMPKAEVKI--GENNEIL---------VRGGMVMKGYYK 432
Query: 533 NPSATNQAIDKDGWLNTGDIG 553
P T QA +DG+L TGD G
Sbjct: 433 KPEETAQAFTEDGFLKTGDAG 453
>LCF6_RAT (P33124) Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase 6) (LACS 6)
(Long-chain-fatty-acid--CoA ligase, brain isozyme)
Length = 697
Score = 105 bits (261), Expect = 5e-22
Identities = 107/422 (25%), Positives = 174/422 (40%), Gaps = 37/422 (8%)
Query: 151 VSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNP 210
V + I +FA N W++A+ + V + I N ++ + VD P
Sbjct: 144 VGTEQFIGVFAQNRPEWIIAELACYTYSMVVVPLYDTLGPGSIRYIINTADICTVIVDKP 203
Query: 211 EMFNRIAKAFDLKAS--MRFVILLWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFES 268
+ + + K + ++ VIL+ L G K + S + E
Sbjct: 204 HKAILLLEHVERKETPGLKLVILMEPFDDALRERGKK----------CGVDIKSMQAIED 253
Query: 269 HDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQIS--------KYAASLA 320
H V + DD++ + +TSGTTGNPKG MLTH N++ S ++A + A
Sbjct: 254 SGQENHRVPVPPRPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTESQWAPTCA 313
Query: 321 CI*KS-L*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLG 379
+ S L + H F R S+ C V + G R +S + + L
Sbjct: 314 DVHFSYLPLAHMFERMVQSVVYCHGGRV--GFFQGDIRLLSDDMKALRPTIFPVVPRLLN 371
Query: 380 YMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPI---HMLAKKLVYSKIHSA 436
M K ++ K +L++ R A + I + + +L ++KI ++
Sbjct: 372 RMYDKIFHQADTSLKRW--------LLEFAAKRKQAEVRSGIIRNNSIWDELFFNKIQAS 423
Query: 437 IGFSKAGISGGGS--LPSHVDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHP 494
+G I G + P+ + A+G + GYG TE + G VG P
Sbjct: 424 LGGHVRMIVTGAAPASPTVLGFLRAALGCQVYEGYGQTECTAGCTFTTPGDWTSGHVGAP 483
Query: 495 LKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIG- 553
L K+VD+E +G + V+GP + KGY K+ T +A+D DGWL+TGDIG
Sbjct: 484 LPCNHIKLVDAEELNYWTSKGEGEICVKGPNVFKGYLKDEDRTKEALDSDGWLHTGDIGK 543
Query: 554 WI 555
W+
Sbjct: 544 WL 545
>4CL2_TOBAC (O24146) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 542
Score = 104 bits (259), Expect = 8e-22
Identities = 113/440 (25%), Positives = 185/440 (41%), Gaps = 84/440 (19%)
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
TY +E A GL G+ P + I + NS ++ A G GAI+ + +
Sbjct: 54 TYADVELNSRKVAAGLHKQGIQPKDTIMILLPNSPEFVFAFIGASYLGAISTMANPLFTP 113
Query: 191 EELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIF 250
E+++ S S + V N++ K + + ++ + + + CL +
Sbjct: 114 AEVVKQAKAS-SAKIIVTQACHVNKV-KDYAFENDVKIICIDSAPEGCL------HFSVL 165
Query: 251 TFTEIMHLGRGSRRLFESHDARKHYVFEA-IKSDDIATLVYTSGTTGNPKGVMLTHQNLL 309
T A +H + E I+ DD+ L Y+SGTTG PKGVMLTH+ L+
Sbjct: 166 T------------------QANEHDIPEVEIQPDDVVALPYSSGTTGLPKGVMLTHKGLV 207
Query: 310 HQISK---------YAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQIS 360
+++ Y S + L +FH IY+ + L G+ R +
Sbjct: 208 TSVAQQVDGENPNLYIHSEDVMLCVLPLFH--------IYSLNSV-----LLCGL-RVGA 253
Query: 361 TSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFP 420
+++K ++F+ + Y + + KSP M+D + T++
Sbjct: 254 AILIMQKFDIVSFLELIQRYKVTIGPFVPPIVLAIAKSP-----MVDDYDLSSVRTVMSG 308
Query: 421 IHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIA- 479
L K+L + + +K G GYG+TE PV+A
Sbjct: 309 AAPLGKEL-EDTVRAKFPNAKLG-----------------------QGYGMTEAGPVLAM 344
Query: 480 ----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPS 535
A+ G+ G +++ E K+VD +TG LP G + +RG Q+MKGY +P
Sbjct: 345 CLAFAKEPFEIKSGACGTVVRNAEMKIVDPKTGNSLPRNQSGEICIRGDQIMKGYLNDPE 404
Query: 536 ATNQAIDKDGWLNTGDIGWI 555
AT + IDK+GWL TGDIG+I
Sbjct: 405 ATARTIDKEGWLYTGDIGYI 424
>4CL_PINTA (P41636) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL)
(4-coumaroyl-CoA synthase)
Length = 537
Score = 101 bits (251), Expect = 7e-21
Identities = 122/465 (26%), Positives = 186/465 (39%), Gaps = 98/465 (21%)
Query: 112 KYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVAD 171
++ D+ LID D T + ++E A GL +G+ + + L N +
Sbjct: 37 EFADRPCLIDGATD--RTYCFSEVELISRKVAAGLAKLGLQQGQVVMLLLPNCIEFAFVF 94
Query: 172 QGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVIL 231
G GAI + A P + AKA A R ++
Sbjct: 95 MGASVRGAI--------------------VTTANPFYKPGEIAKQAKA----AGARIIVT 130
Query: 232 LWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYT 291
L L + S +V + T + G + D + + I DD+ L Y+
Sbjct: 131 LAAYVEKLADLQSHDVLVITIDDAPKEGCQHISVLTEADETQCPAVK-IHPDDVVALPYS 189
Query: 292 SGTTGNPKGVMLTHQNLLHQISK---------YAASLACI*KSL*VFHFFMRC*PSIYNC 342
SGTTG PKGVMLTH+ L+ +++ Y S I L +FH +
Sbjct: 190 SGTTGLPKGVMLTHKGLVSSVAQQVDGENPNLYFHSDDVILCVLPLFHIY---------- 239
Query: 343 EKLEVYESLYSGIQRQISTSSLVRKLVALTFIRVSLGYMECKRIYEGKCLTKNQKSPSYL 402
SL S V L +R + ++ CL QK
Sbjct: 240 -------SLNS---------------VLLCALRAGAATLIMQKFNLTTCLELIQK----- 272
Query: 403 YAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLP-------SHVD 455
Y + +A I+ PI + K S I S S I G+ P + +
Sbjct: 273 YKVT-------VAPIVPPIVLDITK---SPIVSQYDVSSVRIIMSGAAPLGKELEDALRE 322
Query: 456 RFFEAIGVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEV 510
RF +AI GYG+TE PV+A A+ GS G +++ + K++D+ETGE
Sbjct: 323 RFPKAI---FGQGYGMTEAGPVLAMNLAFAKNPFPVKSGSCGTVVRNAQIKILDTETGES 379
Query: 511 LPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
LP G + +RGP++MKGY +P +T ID++GWL+TGD+ +I
Sbjct: 380 LPHNQAGEICIRGPEIMKGYINDPESTAATIDEEGWLHTGDVEYI 424
>LUCI_LUCLA (Q01158) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 548
Score = 96.7 bits (239), Expect = 2e-19
Identities = 78/281 (27%), Positives = 129/281 (45%), Gaps = 44/281 (15%)
Query: 278 EAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*VFHFFMRC*P 337
E + + +A ++ +SG+TG PKGV LTH+N + + S
Sbjct: 187 EVNRKEQVALIMNSSGSTGLPKGVQLTHENAVTRFS------------------------ 222
Query: 338 SIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTF---IRVSLGYMECKRIYEGKCLTK 394
+ +Y Q+S + + +V + +LGY+ C + LTK
Sbjct: 223 --------HARDPIYGN---QVSPGTAILTVVPFHHGFGMFTTLGYLTCG--FRIVMLTK 269
Query: 395 NQKSPSYLYAMLDW-LGARIIATILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSH 453
+ ++L + D+ + I+ LF I ++ L + + + + G +
Sbjct: 270 FDEE-TFLKTLQDYKCSSVILVPTLFAILNRSELLDKYDLSNLVEIASGGAPLSKEIGEA 328
Query: 454 VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPP 513
V R F GV + GYGLTET+ I + G+ G + + KV+D +T + L P
Sbjct: 329 VARRFNLPGV--RQGYGLTETTSAIIITPEGDDKPGASGKVVPLFKAKVIDLDTKKTLGP 386
Query: 514 GYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
+G + V+GP LMKGY NP AT + ID++GWL+TGDIG+
Sbjct: 387 NRRGEVCVKGPMLMKGYVDNPEATREIIDEEGWLHTGDIGY 427
>LUCI_PHOPY (P08659) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 550
Score = 96.3 bits (238), Expect = 2e-19
Identities = 114/438 (26%), Positives = 182/438 (41%), Gaps = 76/438 (17%)
Query: 130 MTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSS 189
+TY + + + AE ++ G++ N +I + ++NS ++ + G + G +
Sbjct: 51 ITYAEYFEMSVRLAEAMKRYGLNTNHRIVVCSENSLQFFMPVLGALFIGVAVAPANDIYN 110
Query: 190 IEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPI 249
ELL N S+ + V + + L + +I+ S +G + +
Sbjct: 111 ERELLNSMNISQPTVVFVSKKGLQKILNVQKKLPIIQKIIIM----DSKTDYQGFQSMYT 166
Query: 250 FTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDD-IATLVYTSGTTGNPKGVMLTHQNL 308
F + HL G F +D +V E+ D IA ++ +SG+TG PKGV L H+
Sbjct: 167 FVTS---HLPPG----FNEYD----FVPESFDRDKTIALIMNSSGSTGLPKGVALPHRT- 214
Query: 309 LHQISKYAASLACI*KSL*VFHFFMRC*PSIYNCEKLEVYESLYSGIQRQISTSSLVRKL 368
AC+ F I+ QI + + +
Sbjct: 215 -----------ACV--------RFSHARDPIFG---------------NQIIPDTAILSV 240
Query: 369 VALTF---IRVSLGYMECK-RIYEGKCLTKNQKSPSYLYAMLDWL--GARIIATIL--FP 420
V + +LGY+ C R+ L + +L ++ D+ A ++ T+ F
Sbjct: 241 VPFHHGFGMFTTLGYLICGFRVV----LMYRFEEELFLRSLQDYKIQSALLVPTLFSFFA 296
Query: 421 IHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSHVD----RFFEAIGVTLQNGYGLTETSP 476
L K S +H SGG L V + F G+ + GYGLTET+
Sbjct: 297 KSTLIDKYDLSNLHEIA-------SGGAPLSKEVGEAVAKRFHLPGI--RQGYGLTETTS 347
Query: 477 VIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSA 536
I + G+VG + E KVVD +TG+ L +G L VRGP +M GY NP A
Sbjct: 348 AILITPEGDDKPGAVGKVVPFFEAKVVDLDTGKTLGVNQRGELCVRGPMIMSGYVNNPEA 407
Query: 537 TNQAIDKDGWLNTGDIGW 554
TN IDKDGWL++GDI +
Sbjct: 408 TNALIDKDGWLHSGDIAY 425
>LUCI_LUCCR (P13129) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 548
Score = 95.1 bits (235), Expect = 5e-19
Identities = 79/281 (28%), Positives = 128/281 (45%), Gaps = 44/281 (15%)
Query: 278 EAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAASLACI*KSL*VFHFFMRC*P 337
E + + +A ++ +SG+TG PKGV LTH+N + + S
Sbjct: 187 EVDRKEQVALIMNSSGSTGLPKGVQLTHENTVTRFS------------------------ 222
Query: 338 SIYNCEKLEVYESLYSGIQRQISTSSLVRKLVALTF---IRVSLGYMECKRIYEGKCLTK 394
+ +Y Q+S + V +V + +LGY+ C + LTK
Sbjct: 223 --------HARDPIYGN---QVSPGTAVLTVVPFHHGFGMFTTLGYLICG--FRVVMLTK 269
Query: 395 NQKSPSYLYAMLDWLGARIIAT-ILFPIHMLAKKLVYSKIHSAIGFSKAGISGGGSLPSH 453
+ ++L + D+ +I LF I ++ L + + + + G +
Sbjct: 270 FDEE-TFLKTLQDYKCTSVILVPTLFAILNKSELLNKYDLSNLVEIASGGAPLSKEVGEA 328
Query: 454 VDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPP 513
V R F GV + GYGLTET+ I + G+ G + + KV+D +T + L P
Sbjct: 329 VARRFNLPGV--RQGYGLTETTSAIIITPEGDDKPGASGKVVPLFKAKVIDLDTKKSLGP 386
Query: 514 GYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGW 554
+G + V+GP LMKGY NP AT + ID++GWL+TGDIG+
Sbjct: 387 NRRGEVCVKGPMLMKGYVNNPEATKELIDEEGWLHTGDIGY 427
>4CL1_ARATH (Q42524) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 561
Score = 87.8 bits (216), Expect = 8e-17
Identities = 82/295 (27%), Positives = 132/295 (43%), Gaps = 67/295 (22%)
Query: 280 IKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK---------YAASLACI*KSL*VFH 330
I DD+ L Y+SGTTG PKGVMLTH+ L+ +++ Y S I L +FH
Sbjct: 199 ISPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFHSDDVILCVLPMFH 258
Query: 331 FF---------MRC*PSIYNCEKLEVYESLYSGIQR-QISTSSLVRKLVALTFIRVSLGY 380
+ +R +I K E+ L IQR +++ + +V +V
Sbjct: 259 IYALNSIMLCGLRVGAAILIMPKFEI-NLLLELIQRCKVTVAPMVPPIVL---------- 307
Query: 381 MECKRIYEGKCLTKNQKSPSYLYAMLDWLGARIIATILFPIHMLAKKLVYSKIHSAIGFS 440
+ K+ ++ Y D R++ + P+ + V +K +A
Sbjct: 308 ----------AIAKSSETEKY-----DLSSIRVVKSGAAPLGKELEDAVNAKFPNAKLGQ 352
Query: 441 KAGISGGGSLPSHVDRFFEAIGVTLQNGYGLTETSPVIAARRLSCNVIGSVGHPLKHTEF 500
G++ G + + + G+ E PV + +C + +++ E
Sbjct: 353 GYGMTEAGPV------------LAMSLGFA-KEPFPVKSG---ACGTV------VRNAEM 390
Query: 501 KVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
K+VD +TG+ L G + +RG Q+MKGY NP+AT + IDKDGWL+TGDIG I
Sbjct: 391 KIVDPDTGDSLSRNQPGEICIRGHQIMKGYLNNPAATAETIDKDGWLHTGDIGLI 445
>LCFA_BACSU (P94547) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 560
Score = 87.4 bits (215), Expect = 1e-16
Identities = 51/124 (41%), Positives = 72/124 (57%), Gaps = 3/124 (2%)
Query: 434 HSAIGFSKAGISGGGSLPSHVDRFFEAI-GVTLQNGYGLTETSPVIAARRL-SCNVIGSV 491
H + K+ +SG +LP V + FE + G L GYGL+E SPV A + N GS+
Sbjct: 319 HYDLSSIKSCLSGSAALPVEVKQKFEKVTGGKLVEGYGLSEASPVTHANFIWGKNKPGSI 378
Query: 492 GHPLKHTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGD 551
G P T+ + ETGE+ P G + V+GPQ+MKGY+ P T A+ +DGWL TGD
Sbjct: 379 GCPWPSTDAAIYSEETGELAAPYEHGEIIVKGPQVMKGYWNKPEET-AAVLRDGWLFTGD 437
Query: 552 IGWI 555
+G++
Sbjct: 438 MGYM 441
Score = 52.4 bits (124), Expect = 4e-06
Identities = 57/232 (24%), Positives = 96/232 (40%), Gaps = 29/232 (12%)
Query: 100 KTVPDIWRSSAEKYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIAL 159
KT+ I SA ++ DK A+ ++ +T+ + L A L+ G+ +++A+
Sbjct: 24 KTLQSILTDSAARFPDKTAI--SFYG--KKLTFHDILTDALKLAAFLQCNGLQKGDRVAV 79
Query: 160 FADNSCRWLVADQGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKA 219
N + +++ G++ G I VV+ + E L+ +++ + +F + K
Sbjct: 80 MLPNCPQTVISYYGVLFAGGI-VVQTNPLYTEHELEYQLRDAQVSVIITLDLLFPKAIKM 138
Query: 220 FDLKASMRFVI------------LLWG--EKSCLVNEGSKEVPIFTFTEIMHLGRGSRRL 265
L + +I +L+ +K + + K I TF M +
Sbjct: 139 KTLSIVDQILITSVKDYLPFPKNILYPLTQKQKVHIDFDKTANIHTFASCMKQEKTELLT 198
Query: 266 FESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISKYAA 317
D DIA L YT GTTG PKGVMLTHQN+L AA
Sbjct: 199 IPKIDPEH----------DIAVLQYTGGTTGAPKGVMLTHQNILANTEMCAA 240
>4CL_VANPL (O24540) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL)
(4-coumaroyl-CoA synthase)
Length = 553
Score = 87.4 bits (215), Expect = 1e-16
Identities = 44/96 (45%), Positives = 61/96 (62%), Gaps = 5/96 (5%)
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+A A+ G+ G +++ E K+VD ETG LP + G +
Sbjct: 338 LGQGYGMTEAGPVLAMCLAFAKEPFDIKSGACGTVVRNAEMKIVDPETGSSLPRNHPGEI 397
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY +P AT + IDK+GWL+TGDIG+I
Sbjct: 398 CIRGDQIMKGYLNDPEATARTIDKEGWLHTGDIGYI 433
Score = 42.4 bits (98), Expect = 0.004
Identities = 50/209 (23%), Positives = 80/209 (37%), Gaps = 34/209 (16%)
Query: 112 KYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVAD 171
K+ + LI+ D TY +E GL +G+ + I + NS ++ A
Sbjct: 41 KFSSRPCLINGATD--EIFTYADVELISRRVGSGLSKLGIKQGDTIMILLPNSPEFVFAF 98
Query: 172 QGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVIL 231
G G+I+ + + E++ + AKA + K +
Sbjct: 99 LGASFIGSISTMANPFFTSTEVI--------------------KQAKASNAKLIITQGCY 138
Query: 232 LWGEKSCLVNEGSKEVPIFTFT------EIMHLGRGSRRLFESHDARKHYVFEAIKSDDI 285
+ K G K + I T T I+H D + E I D +
Sbjct: 139 VDKVKDYACENGVKIISIDTTTTADDAANILHFSE-----LTGADENEMPKVE-ISPDGV 192
Query: 286 ATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
L Y+SGTTG PKGVMLTH+ L+ +++
Sbjct: 193 VALPYSSGTTGLPKGVMLTHKGLVTSVAQ 221
>4CL1_SOYBN (P31686) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1) (Clone 4CL14) (Fragment)
Length = 293
Score = 84.7 bits (208), Expect = 6e-16
Identities = 48/118 (40%), Positives = 65/118 (54%), Gaps = 7/118 (5%)
Query: 445 SGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKH 497
SGG L ++ A L GYG+TE PV+ A+ G+ G +++
Sbjct: 57 SGGAPLGKELEDTLRAKFPNAKLGQGYGMTEAGPVLTMSLAFAKEPIDVKPGACGTVVRN 116
Query: 498 TEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
E K+VD ETG LP G + +RG Q+MKGY + AT + IDKDGWL+TGDIG+I
Sbjct: 117 AEMKIVDPETGHSLPRNQSGEICIRGDQIMKGYLNDGEATERTIDKDGWLHTGDIGYI 174
>4CL1_ORYSA (P17814) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 563
Score = 84.7 bits (208), Expect = 6e-16
Identities = 44/119 (36%), Positives = 68/119 (56%), Gaps = 7/119 (5%)
Query: 444 ISGGGSLPSHVDRFFEAI--GVTLQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLK 496
+SG + ++ F A G L GYG+TE PV++ A+ G+ G ++
Sbjct: 326 LSGAAPMGKDIEDAFMAKLPGAVLGQGYGMTEAGPVLSMCLAFAKEPFKVKSGACGTVVR 385
Query: 497 HTEFKVVDSETGEVLPPGYKGILKVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+ E K++D +TG+ L +G + +RG Q+MKGY NP AT ID +GWL+TGDIG++
Sbjct: 386 NAELKIIDPDTGKSLGRNLRGEICIRGQQIMKGYLNNPEATKNTIDAEGWLHTGDIGYV 444
Score = 42.7 bits (99), Expect = 0.003
Identities = 18/32 (56%), Positives = 25/32 (77%)
Query: 283 DDIATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
DD+ L Y+SGTTG PKGVMLTH++L +++
Sbjct: 201 DDVVALPYSSGTTGLPKGVMLTHRSLSTSVAQ 232
>4CL1_TOBAC (O24145) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 547
Score = 84.0 bits (206), Expect = 1e-15
Identities = 43/96 (44%), Positives = 60/96 (61%), Gaps = 5/96 (5%)
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+A A+ G+ G +++ E K+VD +TG LP G +
Sbjct: 334 LGQGYGMTEAGPVLAMCLAFAKEPFDIKSGACGTVVRNAEMKIVDPDTGCSLPRNQPGEI 393
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY +P AT + IDK+GWL+TGDIG+I
Sbjct: 394 CIRGDQIMKGYLNDPEATTRTIDKEGWLHTGDIGFI 429
Score = 52.0 bits (123), Expect = 5e-06
Identities = 45/185 (24%), Positives = 80/185 (42%), Gaps = 27/185 (14%)
Query: 131 TYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVADQGMMATGAINVVRGSRSSI 190
TY ++E A GL +G+ + I + NS ++ A G GAI+ + +
Sbjct: 59 TYAEVELTCRKVAVGLNKLGIQQKDTIMILLPNSPEFVFAFMGASYLGAISTMANPLFTP 118
Query: 191 EELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVILLWGEKSCLVNEGSKEVPIF 250
E+++ S A + F K + + ++ + + + CL
Sbjct: 119 AEVVK--QAKASSAKIIITQSCFVGKVKDYASENDVKVICIDSAPEGCL----------- 165
Query: 251 TFTEIMHLGRGSRRLFESHDARKHYVFEA-IKSDDIATLVYTSGTTGNPKGVMLTHQNLL 309
F+E+ + +H + E I+ DD+ L Y+SGTTG PKGVMLTH+ L+
Sbjct: 166 HFSELTQ-------------SDEHEIPEVKIQPDDVVALPYSSGTTGLPKGVMLTHKGLV 212
Query: 310 HQISK 314
+++
Sbjct: 213 TSVAQ 217
>4CL2_ARATH (Q9S725) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 556
Score = 83.6 bits (205), Expect = 1e-15
Identities = 42/96 (43%), Positives = 60/96 (61%), Gaps = 5/96 (5%)
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+A A+ G+ G +++ E K++D +TG+ LP G +
Sbjct: 343 LGQGYGMTEAGPVLAMSLGFAKEPFPVKSGACGTVVRNAEMKILDPDTGDSLPRNKPGEI 402
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY +P AT IDKDGWL+TGD+G+I
Sbjct: 403 CIRGNQIMKGYLNDPLATASTIDKDGWLHTGDVGFI 438
Score = 46.2 bits (108), Expect = 3e-04
Identities = 21/50 (42%), Positives = 33/50 (66%)
Query: 265 LFESHDARKHYVFEAIKSDDIATLVYTSGTTGNPKGVMLTHQNLLHQISK 314
L +S + R + E I +D+ L ++SGTTG PKGVMLTH+ L+ +++
Sbjct: 177 LTQSEEPRVDSIPEKISPEDVVALPFSSGTTGLPKGVMLTHKGLVTSVAQ 226
>4CL2_SOLTU (P31685) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 545
Score = 81.6 bits (200), Expect = 5e-15
Identities = 42/96 (43%), Positives = 60/96 (61%), Gaps = 5/96 (5%)
Query: 465 LQNGYGLTETSPVIA-----ARRLSCNVIGSVGHPLKHTEFKVVDSETGEVLPPGYKGIL 519
L GYG+TE PV+A A+ G+ G +++ E K+VD +TG LP G +
Sbjct: 332 LGQGYGMTEAGPVLAMCLAFAKEPFDIKSGACGTVVRNAEMKIVDPDTGCSLPRNQPGEI 391
Query: 520 KVRGPQLMKGYYKNPSATNQAIDKDGWLNTGDIGWI 555
+RG Q+MKGY +P AT + I+K+GWL+TGDIG+I
Sbjct: 392 CIRGDQIMKGYLNDPEATARTIEKEGWLHTGDIGFI 427
Score = 50.4 bits (119), Expect = 1e-05
Identities = 48/203 (23%), Positives = 87/203 (42%), Gaps = 27/203 (13%)
Query: 112 KYGDKIALIDPYHDPPSTMTYKQLEDAILDFAEGLRVIGVSPNEKIALFADNSCRWLVAD 171
++ + LID +D TY ++E A GL +G+ + I + N ++ A
Sbjct: 40 EFNSRPCLIDGAND--RIYTYAEVELTSRKVAVGLNKLGIQQKDTIMILLPNCPEFVFAF 97
Query: 172 QGMMATGAINVVRGSRSSIEELLQIYNHSESIALAVDNPEMFNRIAKAFDLKASMRFVIL 231
G GAI+ + + E+++ S A V F K + ++ ++ + +
Sbjct: 98 IGASYLGAISTMANPLFTPAEVVK--QAKASSAKIVITQACFAGKVKDYAIENDLKVICV 155
Query: 232 LWGEKSCLVNEGSKEVPIFTFTEIMHLGRGSRRLFESHDARKHYVFEAIKSDDIATLVYT 291
+ C+ F+E++ E D + I+ DD+ L Y+
Sbjct: 156 DSAPEGCV-----------HFSELIQSDE-----HEIPDVK-------IQPDDVVALPYS 192
Query: 292 SGTTGNPKGVMLTHQNLLHQISK 314
SGTTG PKGVMLTH+ L+ +++
Sbjct: 193 SGTTGLPKGVMLTHKGLVTSVAQ 215
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,313,141
Number of Sequences: 164201
Number of extensions: 2985570
Number of successful extensions: 9258
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 187
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 8603
Number of HSP's gapped (non-prelim): 544
length of query: 617
length of database: 59,974,054
effective HSP length: 116
effective length of query: 501
effective length of database: 40,926,738
effective search space: 20504295738
effective search space used: 20504295738
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146706.7


