

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146683.4 + phase: 0 /pseudo
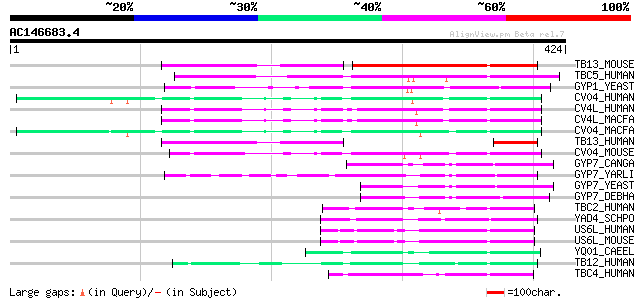
(424 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TB13_MOUSE (Q8R3D1) TBC1 domain family member 13 146 9e-35
TBC5_HUMAN (Q92609) TBC1 domain family member 5 124 5e-28
GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1) 105 2e-22
CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4 100 7e-21
CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197 100 9e-21
CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog ... 100 1e-20
CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog (... 99 2e-20
TB13_HUMAN (Q9NVG8) TBC1 domain family member 13 96 1e-19
CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog (... 96 1e-19
GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7) 71 6e-12
GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7) 64 7e-10
GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7) 61 5e-09
GYP7_DEBHA (Q6BU76) GTPase-activating protein GYP7 (GAP for YPT7) 60 8e-09
TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antige... 53 1e-06
YAD4_SCHPO (Q09830) Hypothetical protein C4G8.04 in chromosome I 51 6e-06
US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the... 50 8e-06
US6L_MOUSE (Q80XC3) USP6 N-terminal like protein 50 1e-05
YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II 50 1e-05
TB12_HUMAN (O60347) TBC1 domain family member 12 49 2e-05
TBC4_HUMAN (O60343) TBC1 domain family member 4 47 1e-04
>TB13_MOUSE (Q8R3D1) TBC1 domain family member 13
Length = 400
Score = 146 bits (369), Expect = 9e-35
Identities = 70/141 (49%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Query: 263 KSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVE 322
+++ E ++ IL I+AKLNPGI YVQGMNE++ PL+Y F DP+ E +EADTFFCF
Sbjct: 205 EAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDPNSEWKEHAEADTFFCFTN 264
Query: 323 LLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITL 382
L++ RDNF + LD+S GI + ++ LK+ D EL+ L+ + PQF+AFRW+TL
Sbjct: 265 LMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKLQ-EQSIKPQFFAFRWLTL 323
Query: 383 LLTQEFDFADSLRIWDTLVSD 403
LL+QEF D +RIWD+L +D
Sbjct: 324 LLSQEFLLPDVIRIWDSLFAD 344
Score = 94.0 bits (232), Expect = 7e-19
Identities = 49/139 (35%), Positives = 76/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPSIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRGLYSQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M + R +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVFREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
>TBC5_HUMAN (Q92609) TBC1 domain family member 5
Length = 795
Score = 124 bits (311), Expect = 5e-28
Identities = 93/333 (27%), Positives = 145/333 (42%), Gaps = 68/333 (20%)
Query: 127 LRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILINPSEIT 186
+R+ G S RS WKL L LP D+S W S + + R+ Y K+ + NP ++
Sbjct: 72 IRQKGINGQLRSSRFRSICWKLFLCVLPQDKSQWISRIEELRAWYSNIKEIHITNPRKVV 131
Query: 187 RRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDRDVK 246
+ Q +PLS + S+WN+FFQD ++ I++DVK
Sbjct: 132 GQ-----------------------QDLMINNPLSQDEGSLWNKFFQDKELRSMIEQDVK 168
Query: 247 RTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND--- 303
RT P+M FF ++ ++ L ++L +A+ N + Y QGM+E+LAP+ +V D
Sbjct: 169 RTFPEMQFF-----QQENVRKILTDVLFCYARENEQLLYKQGMHELLAPIVFVLHCDHQA 223
Query: 304 ---------PDEE-----NAAFSEADTFFCFVELLSGFRDNFC----------------- 332
P EE N + E D + F +L+ F
Sbjct: 224 FLHASESAQPSEEMKTVLNPEYLEHDAYAVFSQLMETAEPWFSTFEHDGQKGKETLMTPI 283
Query: 333 -----QQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQE 387
Q L +I + LLK+HD EL+ HL ++ PQ Y RW+ LL +E
Sbjct: 284 PFARPQDLGPTIAIVTKVNQIQDHLLKKHDIELYMHLN-RLEIAPQIYGLRWVRLLFGRE 342
Query: 388 FDFADSLRIWDTLVSDPDGPQVQIYSFLPCIVF 420
F D L +WD L +D + Y F+ +++
Sbjct: 343 FPLQDLLVVWDALFADGLSLGLVDYIFVAMLLY 375
>GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1)
Length = 637
Score = 105 bits (262), Expect = 2e-22
Identities = 88/313 (28%), Positives = 134/313 (42%), Gaps = 78/313 (24%)
Query: 119 RKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDI 178
+ +I+ ++LR+I+ GIP R +WKLL+GYLP + L +KR +Y+
Sbjct: 265 KTIINQQDLRQISWNGIPKIH--RPVVWKLLIGYLPVNTKRQEGFLQRKRKEYR------ 316
Query: 179 LINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDII 238
D +K H S +Q +D
Sbjct: 317 -------------------DSLK----------------HTFS-------DQHSRDIPTW 334
Query: 239 EQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFY 298
QI+ D+ RT+P + + KS Q +L+ IL ++A +P YVQG+N+++ P F
Sbjct: 335 HQIEIDIPRTNPHIPLY-----QFKSVQNSLQRILYLWAIRHPASGYVQGINDLVTPFFE 389
Query: 299 VFKN--------------DPD----EENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIV 340
F DP +E EADTF+C +LL DN+ +
Sbjct: 390 TFLTEYLPPSQIDDVEIKDPSTYMVDEQITDLEADTFWCLTKLLEQITDNYI----HGQP 445
Query: 341 GIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
GI + LSQL+K D +L+ H + QF AFRW+ LL +EF +R+WDT
Sbjct: 446 GILRQVKNLSQLVKRIDADLYNHFQNEHVEFIQF-AFRWMNCLLMREFQMGTVIRMWDTY 504
Query: 401 VSDPDGPQVQIYS 413
+S+ YS
Sbjct: 505 LSETSQEVTSSYS 517
>CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4
Length = 517
Score = 100 bits (249), Expect = 7e-21
Identities = 113/420 (26%), Positives = 171/420 (39%), Gaps = 84/420 (20%)
Query: 6 VPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAS 65
V + N + + Q R + P P P PPS + S S +S +
Sbjct: 94 VMETANRVLRNHSQRQGRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPA 153
Query: 66 TSSTDDMSISR---LAQSQSQLLAEV--PFTLKSKCL*LLFSIQLY*YC*LFVY**LSRK 120
S++D + R L S + L P TL S L + +L + L L+
Sbjct: 154 ESASDAAPLQRSQSLPHSATVTLGGTSDPSTLSSSALSEREASRLDKFKQL-----LAGP 208
Query: 121 VIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILI 180
D+ ELR+++ GIP +R WKLL GYLP + + L +K+ +Y F +
Sbjct: 209 NTDLEELRRLSWSGIPKP--VRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHY-- 264
Query: 181 NPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQ 240
YD SR+ H QDT Q
Sbjct: 265 --------------YD------------SRNDEVH-----------------QDT--YRQ 279
Query: 241 IDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVF 300
I D+ R P+ L E + IL I+A +P YVQG+N+++ P F VF
Sbjct: 280 IHIDIPRMSPEALI------LQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVVF 333
Query: 301 KNDPDE--------------ENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTI 346
+ E E EADT++C +LL G +DN+ + GI+ +
Sbjct: 334 ICEYIEAEEVDTVDVSGVPAEVLCNIEADTYWCMSKLLDGIQDNY----TFAQPGIQMKV 389
Query: 347 TRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDG 406
L +L+ DE++ RHL+ +V +AFRW+ LL +E ++R+WDT S+PDG
Sbjct: 390 KMLEELVSRIDEQVHRHLD-QHEVRYLQFAFRWMNNLLMREVPLRCTIRLWDTYQSEPDG 448
>CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197
Length = 505
Score = 100 bits (248), Expect = 9e-21
Identities = 88/303 (29%), Positives = 134/303 (44%), Gaps = 72/303 (23%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS + D+ ELRK + G+P +R W+LL GYLP + L +KR +Y F +
Sbjct: 193 LSSQNTDLDELRKCSWPGVPRE--VRPITWRLLSGYLPANTERRKLTLQRKREEYFGFIE 250
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
YD SR++ H QDT
Sbjct: 251 QY----------------YD------------SRNEEHH-----------------QDT- 264
Query: 237 IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
QI D+ RT+P + F L QE + IL I+A +P YVQG+N+++ P
Sbjct: 265 -YRQIHIDIPRTNPLIPLF--QQPLV---QEIFERILFIWAIRHPASGYVQGINDLVTPF 318
Query: 297 FYVFKNDPDEENA-------------AFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIR 343
F VF ++ EE+ EAD+F+C +LL G +DN+ + GI+
Sbjct: 319 FVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKLLDGIQDNY----TFAQPGIQ 374
Query: 344 STITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
+ L +L+ DE++ H +V +AFRW+ LL +E ++R+WDT S+
Sbjct: 375 KKVKALEELVSRIDEQVHNHFR-RYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSE 433
Query: 404 PDG 406
P+G
Sbjct: 434 PEG 436
>CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog
(QtsA-20424)
Length = 505
Score = 99.8 bits (247), Expect = 1e-20
Identities = 88/303 (29%), Positives = 133/303 (43%), Gaps = 72/303 (23%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
LS D+ ELRK + G+P +R W+LL GYLP + L +KR +Y F +
Sbjct: 193 LSSHNTDLDELRKCSWPGVPRE--VRPVTWRLLSGYLPANTERRKLTLQRKREEYFGFIE 250
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
YD SR++ H QDT
Sbjct: 251 QY----------------YD------------SRNEEHH-----------------QDT- 264
Query: 237 IIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
QI D+ RT+P + F L QE + IL I+A +P YVQG+N+++ P
Sbjct: 265 -YRQIHIDIPRTNPLIPLF--QQPLV---QEIFERILFIWAIRHPASGYVQGINDLVTPF 318
Query: 297 FYVFKNDPDEENA-------------AFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIR 343
F VF ++ EE+ EAD+F+C +LL G +DN+ + GI+
Sbjct: 319 FVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKLLDGIQDNY----TFAQPGIQ 374
Query: 344 STITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
+ L +L+ DE++ H +V +AFRW+ LL +E ++R+WDT S+
Sbjct: 375 KKVKALEELVSRIDEQVHNHFR-RYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSE 433
Query: 404 PDG 406
P+G
Sbjct: 434 PEG 436
>CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog
(QtrA-11492) (Fragment)
Length = 497
Score = 99.0 bits (245), Expect = 2e-20
Identities = 112/421 (26%), Positives = 170/421 (39%), Gaps = 86/421 (20%)
Query: 6 VPDWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSPPSSSSSSAS 65
V + N + + Q R + P P P PPS + S S +S +
Sbjct: 74 VMETANRVLRNHSQRQGRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPA 133
Query: 66 TSSTDDMSISRLAQSQSQLLAEV------PFTLKSKCL*LLFSIQLY*YC*LFVY**LSR 119
S++D + R +QS A P TL S L + +L + L L+
Sbjct: 134 ESASDAAPLQR-SQSLPHAAAVTLGGTSDPGTLSSSALSEREASRLDKFEQL-----LAG 187
Query: 120 KVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDIL 179
D+ ELRK++ GIP +R WKLL GYLP + + L +K+ +Y F +
Sbjct: 188 PNTDLEELRKLSWSGIPKP--VRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHY- 244
Query: 180 INPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIE 239
YD SR+ H QDT
Sbjct: 245 ---------------YD------------SRNDEVH-----------------QDT--YR 258
Query: 240 QIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYV 299
QI D+ R P+ L E + IL I+A +P YVQG+N+++ P F V
Sbjct: 259 QIHIDIPRMSPEALI------LQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVV 312
Query: 300 FKNDPDEENAAFS--------------EADTFFCFVELLSGFRDNFCQQLDNSIVGIRST 345
F + E + EADT++C +LL G +DN+ GI+
Sbjct: 313 FICEYIEAEEVDTVDVSGVPAEVLRNIEADTYWCMSKLLDGIQDNYTFAQP----GIQMK 368
Query: 346 ITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPD 405
+ L +L+ DE++ RHL+ +V +AFRW+ LL +E ++R+WDT S+P+
Sbjct: 369 VKMLEELVSRIDEQVHRHLD-QHEVRYLQFAFRWMNNLLMREVPLRCTIRLWDTYQSEPE 427
Query: 406 G 406
G
Sbjct: 428 G 428
>TB13_HUMAN (Q9NVG8) TBC1 domain family member 13
Length = 275
Score = 96.3 bits (238), Expect = 1e-19
Identities = 50/139 (35%), Positives = 77/139 (54%), Gaps = 17/139 (12%)
Query: 117 LSRKVIDMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQ 176
L I + +LR+++ GIP GLR WK+LL YLP +R+ W+S LAK+R Y +F +
Sbjct: 16 LKEPSIALEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQRELYAQFLR 75
Query: 177 DILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTD 236
+++I P M +SR +T +HPL+ S WN +F+D +
Sbjct: 76 EMIIQPGIAKANM-----------------GVSREDVTFEDHPLNPNPDSRWNTYFKDNE 118
Query: 237 IIEQIDRDVKRTHPDMHFF 255
++ QID+DV+R PD+ FF
Sbjct: 119 VLLQIDKDVRRLCPDISFF 137
Score = 51.2 bits (121), Expect = 5e-06
Identities = 21/34 (61%), Positives = 27/34 (78%)
Query: 370 VNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
+ PQF+AFRW+TLLL+QEF D RIWD+L +D
Sbjct: 186 IKPQFFAFRWLTLLLSQEFLLPDVTRIWDSLFAD 219
>CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog
(Fragment)
Length = 353
Score = 96.3 bits (238), Expect = 1e-19
Identities = 84/298 (28%), Positives = 128/298 (42%), Gaps = 74/298 (24%)
Query: 123 DMRELRKIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQDILINP 182
D+ ELRK++ GIP +R WKLL GYLP + + L +K+ +Y F +
Sbjct: 47 DLEELRKLSWSGIPKP--VRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHYYS-- 102
Query: 183 SEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQID 242
SR+ H QDT QI
Sbjct: 103 --------------------------SRNDEVH-----------------QDT--YRQIH 117
Query: 243 RDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVF-- 300
D+ R P+ L E + IL I+A +P YVQG+N+++ P F VF
Sbjct: 118 IDIPRMSPEALI------LQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVVFIC 171
Query: 301 ----KNDPDEENAAFS--------EADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITR 348
+ D D+ + + EADT++C +LL G +DN+ GI+ +
Sbjct: 172 EYTDREDVDKVDVSSVPAEVLRNIEADTYWCMSKLLDGIQDNYTFAQP----GIQMKVKM 227
Query: 349 LSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPDG 406
L +L+ DE + RHL+ +V +AFRW+ LL +E ++R+WDT S+P+G
Sbjct: 228 LEELVSRIDERVHRHLD-GHEVRYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEG 284
>GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 745
Score = 70.9 bits (172), Expect = 6e-12
Identities = 47/158 (29%), Positives = 76/158 (47%), Gaps = 15/158 (9%)
Query: 258 DSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTF 317
D + N + L +IL + NP + YVQGM ++L+PL+Y+ + DEE TF
Sbjct: 492 DDSIKNPNLKKLADILTTYNIFNPNLGYVQGMTDLLSPLYYIIR---DEET-------TF 541
Query: 318 FCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAF 377
+CF + NF + D S GIR + L+ L + L HL+ + F+ F
Sbjct: 542 WCFTNFMERMERNFLR--DQS--GIRDQMLALTDLCQLMLPRLSAHLQKCDS-SDLFFCF 596
Query: 378 RWITLLLTQEFDFADSLRIWDTLVSDPDGPQVQIYSFL 415
R + + +EF++ D IW+ +D Q Q++ L
Sbjct: 597 RMLLVWFKREFNYDDIFNIWEVFFTDFYSSQYQLFFML 634
>GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 730
Score = 63.9 bits (154), Expect = 7e-10
Identities = 70/287 (24%), Positives = 130/287 (44%), Gaps = 42/287 (14%)
Query: 119 RKVIDMRELR-KIASQGIPDSPGLRSTIWKLLLGYLPPDRSLWSSELAKKRSQYKRFKQD 177
R ++ + E++ +I G+ +P +R W LLG P W S A+++ + + D
Sbjct: 372 RLIVTVNEVKERIFHGGL--APAVRPEGWLFLLGVYP-----WDSTAAERKELVSKLRVD 424
Query: 178 ILINPSEITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDI 237
+ + + + + DD R LSR + + +T FF + D
Sbjct: 425 Y----NRLKKEWWVQEDKERDDF---WRDQLSRIE-------KDVHRTDRNITFFAECDA 470
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKS-NQEALKNILIIFAKLNPGIRYVQGMNEVLAPL 296
+ D D + D F SQ+ + + L+++LI + + N + YVQGM+++L+PL
Sbjct: 471 KKDGDDD----NYDKDEFGFSSQINSNIHLIQLRDMLITYNQHNKNLGYVQGMSDLLSPL 526
Query: 297 FYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEH 356
+ V ++D F+ F + N+ + D S G+R+ + L L++
Sbjct: 527 YVVLQDD----------TLAFWAFSAFMERMERNYLR--DQS--GMRNQLLCLDHLVQFM 572
Query: 357 DEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
L++HLE T N F+ FR + + +E + D LR+W+ L +D
Sbjct: 573 LPSLYKHLEKTESTN-LFFFFRMLLVWFKRELLWDDVLRLWEVLWTD 618
>GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 746
Score = 61.2 bits (147), Expect = 5e-09
Identities = 43/147 (29%), Positives = 69/147 (46%), Gaps = 15/147 (10%)
Query: 269 LKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFR 328
L+NILI + N + YVQGM ++L+P++ + K E TF+CF +
Sbjct: 513 LQNILITYNVYNTNLGYVQGMTDLLSPIYVIMK----------EEWKTFWCFTHFMDIME 562
Query: 329 DNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEF 388
NF + D S GI + L +L++ EL HL N F+ FR + + +EF
Sbjct: 563 RNFLR--DQS--GIHEQMLTLVELVQLMLPELSEHLNKCDSGN-LFFCFRMLLVWFKREF 617
Query: 389 DFADSLRIWDTLVSDPDGPQVQIYSFL 415
+ D + IW+ + Q Q++ L
Sbjct: 618 EMEDIMHIWENFWTFYYSSQFQLFFML 644
>GYP7_DEBHA (Q6BU76) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 757
Score = 60.5 bits (145), Expect = 8e-09
Identities = 38/144 (26%), Positives = 78/144 (53%), Gaps = 15/144 (10%)
Query: 269 LKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFR 328
++ IL+ + + N + YVQGM ++L+PL+ F++ E+ TF+ FV +
Sbjct: 520 MREILLTYNEHNVNLGYVQGMTDLLSPLYVTFQD----------ESLTFWAFVNFMDRME 569
Query: 329 DNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEF 388
NF + D S G+++ + L++L++ +L++HLE + F+ FR + + +EF
Sbjct: 570 RNFLR--DQS--GMKNQMLTLNELVQFMLPDLFKHLEKCESTDLYFF-FRMLLVWFKREF 624
Query: 389 DFADSLRIWDTLVSDPDGPQVQIY 412
+++ L +W+ L +D Q ++
Sbjct: 625 EWSSVLSLWEILWTDYYSGQFHLF 648
>TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antigen
recognized and indentified by SEREX) (PARIS-1)
Length = 917
Score = 53.1 bits (126), Expect = 1e-06
Identities = 49/165 (29%), Positives = 71/165 (42%), Gaps = 22/165 (13%)
Query: 240 QIDRDVKRTHPD-MHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFY 298
QI+ D+ RT P+ HF C S S + L+ +L+ F+ NP I Y QG+N + A
Sbjct: 669 QIELDLNRTFPNNKHFTCPTS----SFPDKLRRVLLAFSWQNPTIGYCQGLNRLAAIALL 724
Query: 299 VFKNDPDEENAAFSEADTFFCFVELLSGF--RDNFCQQLDNSIVGIRSTITRLSQLLKEH 356
V + +EE+A F+C V ++ D +C L S V R L LL E
Sbjct: 725 VLE---EEESA-------FWCLVAIVETIMPADYYCNTLTASQVDQRV----LQDLLSEK 770
Query: 357 DEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLV 401
L HL V+ F W ++ LR+WD +
Sbjct: 771 LPRLMAHLG-QHHVDLSLVTFNWFLVVFADSLISNILLRVWDAFL 814
>YAD4_SCHPO (Q09830) Hypothetical protein C4G8.04 in chromosome I
Length = 772
Score = 50.8 bits (120), Expect = 6e-06
Identities = 44/166 (26%), Positives = 70/166 (41%), Gaps = 17/166 (10%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
+ QID D+ RT FF G L+ IL+ +++ NP I Y QGMN + A L
Sbjct: 544 VAQIDMDINRTMAKNVFFGGKGP----GIPKLRRILVAYSRHNPHIGYCQGMNVIGAFLL 599
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
++ SE D F+ + ++ + ++ R+ L +KE
Sbjct: 600 LLYA----------SEEDAFYMLMSIIENVLPP--KYFTPDLMTSRADQLVLKSFVKESL 647
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
E++ HLE+ V+ +F W + T S RI+D L D
Sbjct: 648 PEIYSHLEL-LGVDLDAISFHWFLSVYTDTLPTNISFRIFDMLFCD 692
>US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the N
terminus of tre) (RN-tre)
Length = 828
Score = 50.4 bits (119), Expect = 8e-06
Identities = 43/164 (26%), Positives = 71/164 (43%), Gaps = 15/164 (9%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
I QID DV RT D H D K Q++L ++L ++ N + Y QGM+++ A L
Sbjct: 141 IRQIDLDVNRTFRD-HIMFRDRYGVK--QQSLFHVLAAYSIYNTEVGYCQGMSQITA-LL 196
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
++ N E D F+ V+L SG + + ++L +
Sbjct: 197 LMYMN----------EEDAFWALVKLFSGPKHAMHGFFVQGFPKLLRFQEHHEKILNKFL 246
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLV 401
+L +HL+ + ++ FY +W F +LRIWD +
Sbjct: 247 SKLKQHLD-SQEIYTSFYTMKWFFQCFLDRTPFTLNLRIWDIYI 289
>US6L_MOUSE (Q80XC3) USP6 N-terminal like protein
Length = 819
Score = 50.1 bits (118), Expect = 1e-05
Identities = 43/164 (26%), Positives = 71/164 (43%), Gaps = 15/164 (9%)
Query: 238 IEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLF 297
I QID DV RT D H D K Q++L ++L ++ N + Y QGM+++ A L
Sbjct: 141 IRQIDLDVNRTFRD-HIMFRDRYGVK--QQSLFHVLAAYSIYNTEVGYCQGMSQITA-LL 196
Query: 298 YVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHD 357
++ N E D F+ V+L SG + + ++L +
Sbjct: 197 LMYMN----------EEDAFWALVKLFSGPKHAMHGFFVQGFPKLLRFQEHHEKILNKFL 246
Query: 358 EELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLV 401
+L +HL+ + ++ FY +W F +LRIWD +
Sbjct: 247 SKLKQHLD-SQEIYTSFYTMKWFFQCFLDRTPFRLNLRIWDIYI 289
>YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II
Length = 330
Score = 49.7 bits (117), Expect = 1e-05
Identities = 42/179 (23%), Positives = 70/179 (38%), Gaps = 20/179 (11%)
Query: 227 IWNQFFQDTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYV 286
+W + ++I+QI D+ RT PD F + ++ L L A+ P + Y
Sbjct: 99 VWQRHEVPDEVIKQIKLDLPRTFPDNKFL-----KTEGTRKTLGRALFAVAEHIPSVGYC 153
Query: 287 QGMNEVLAPLFYVFKNDPDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTI 346
QG+N V + V DE A ++LL + +++G+R +
Sbjct: 154 QGLNFVAGVILLVVN---DESRA-----------IDLLVHLVSQRQEYYGKNMIGLRRDM 199
Query: 347 TRLSQLLKEHDEELWRHLEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSDPD 405
L LL+EH + LE V +W + LR+WD L+ + D
Sbjct: 200 HVLHSLLREHCPRVVVTLE-KLDVGLDMLVGKWFVCWFVESLPMETVLRLWDCLIYEGD 257
>TB12_HUMAN (O60347) TBC1 domain family member 12
Length = 769
Score = 49.3 bits (116), Expect = 2e-05
Identities = 59/280 (21%), Positives = 103/280 (36%), Gaps = 64/280 (22%)
Query: 125 RELRKIASQGIPDSPGLRSTIWKLLLGY-LPPDRSLWSSELAKKRSQYKRFKQDILINPS 183
R +R++ QG+P P +R +W L +G L L+ L++ + ++K F +
Sbjct: 469 RRVRELWWQGLP--PSVRGKVWSLAVGNELNITPELYEIFLSRAKERWKSFSE------- 519
Query: 184 EITRRMFNSASYDADDVKCETRGMLSRSQITHGEHPLSLGKTSIWNQFFQDTDIIEQIDR 243
S+ D + V R +E I
Sbjct: 520 -------TSSENDTEGVSVADREAS-----------------------------LELIKL 543
Query: 244 DVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
D+ RT P ++ F Q + L +IL + P + YVQGM+ + A L +
Sbjct: 544 DISRTFPSLYIF----QKGGPYHDVLHSILGAYTCYRPDVGYVQGMSFIAAVLILNLE-- 597
Query: 304 PDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRH 363
EAD F F LL+ ++D+S++ +E+ +L+ H
Sbjct: 598 ---------EADAFIAFANLLNKPCQLAFFRVDHSMM--LKYFATFEVFFEENLSKLFLH 646
Query: 364 LEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTLVSD 403
+ + + P Y WI L ++ + R+WD D
Sbjct: 647 FK-SYSLTPDIYLIDWIFTLYSKSLPLDLACRVWDVFCRD 685
>TBC4_HUMAN (O60343) TBC1 domain family member 4
Length = 1299
Score = 46.6 bits (109), Expect = 1e-04
Identities = 39/157 (24%), Positives = 69/157 (43%), Gaps = 17/157 (10%)
Query: 244 DVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVLAPLFYVFKND 303
D+ RT P +F S Q +L N+L ++ L+ + Y QG++ V L +
Sbjct: 970 DLGRTFPTHPYF---SVQLGPGQLSLFNLLKAYSLLDKEVGYCQGISFVAGVLLLHMSEE 1026
Query: 304 PDEENAAFSEADTFFCFVELLSGFRDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRH 363
E F D GFR +Q ++ ++ + +LS+LL ++ +L+ H
Sbjct: 1027 QAFEMLKFLMYDL---------GFR----KQYRPDMMSLQIQMYQLSRLLHDYHRDLYNH 1073
Query: 364 LEVTTKVNPQFYAFRWITLLLTQEFDFADSLRIWDTL 400
LE +++P YA W L +F R++D +
Sbjct: 1074 LE-ENEISPSLYAAPWFLTLFASQFSLGFVARVFDII 1109
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.139 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,709,224
Number of Sequences: 164201
Number of extensions: 1951311
Number of successful extensions: 14896
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 103
Number of HSP's that attempted gapping in prelim test: 13816
Number of HSP's gapped (non-prelim): 706
length of query: 424
length of database: 59,974,054
effective HSP length: 113
effective length of query: 311
effective length of database: 41,419,341
effective search space: 12881415051
effective search space used: 12881415051
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146683.4


