

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146681.8 + phase: 0
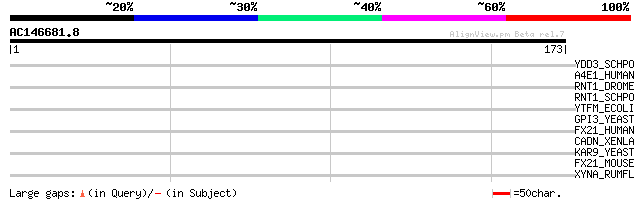
(173 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YDD3_SCHPO (O14206) Hypothetical protein C1B9.03c in chromosome I 33 0.25
A4E1_HUMAN (Q9UPM8) Adapter-related protein complex 4 epsilon 1 ... 31 1.2
RNT1_DROME (Q9VYS3) Regulator of nonsense transcripts 1 homolog 31 1.6
RNT1_SCHPO (Q09820) Regulator of nonsense transcripts 1 homolog 30 2.7
YTFM_ECOLI (P39320) Hypothetical protein ytfM precursor 30 3.6
GPI3_YEAST (P32363) Phosphatidylinositol N-acetylglucosaminyltra... 29 4.7
FX21_HUMAN (O94952) F-box only protein 21 29 4.7
CADN_XENLA (P20310) Neural-cadherin 1 precursor (N-cadherin 1) 29 4.7
KAR9_YEAST (P32526) Karyogamy protein KAR9 (Cortical protein KAR9) 29 6.1
FX21_MOUSE (Q8VDH1) F-box only protein 21 29 6.1
XYNA_RUMFL (P29126) Bifunctional endo-1,4-beta-xylanase xylA pre... 28 8.0
>YDD3_SCHPO (O14206) Hypothetical protein C1B9.03c in chromosome I
Length = 389
Score = 33.5 bits (75), Expect = 0.25
Identities = 18/74 (24%), Positives = 36/74 (48%)
Query: 19 ANQLLFGEDEVIAIIPVIYELPSHLLDDLISRLTPHALYHFHLHMPFQDVNEEDFSRDDS 78
A+QLL + + I +I + + L+ + LYH H+H +++ ++D + S
Sbjct: 276 ASQLLKPKQQAIKLIEIGPRMTLELIKITEDAMGGKVLYHSHVHKSKEEIKQQDNFHEQS 335
Query: 79 TNNKRKRSRDWNLN 92
K KR ++ + N
Sbjct: 336 RALKEKRKKEQDEN 349
>A4E1_HUMAN (Q9UPM8) Adapter-related protein complex 4 epsilon 1
subunit (Epsilon subunit of AP-4) (AP-4 adapter complex
epsilon subunit)
Length = 1137
Score = 31.2 bits (69), Expect = 1.2
Identities = 17/66 (25%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Query: 1 MVEEAPPSSLITLCIDHLANQLLFGEDEVIAIIPVIYELPSHLLDDLISRLTPHALYHFH 60
+V++ ++L+ +C+ +F + + A++P+I + H +++ R ALY FH
Sbjct: 138 VVKDLQSTNLVEVCMALTVVSQIFPREMIPAVLPLIEDKLQH-SKEIVRRKAVLALYKFH 196
Query: 61 LHMPFQ 66
L P Q
Sbjct: 197 LIAPNQ 202
>RNT1_DROME (Q9VYS3) Regulator of nonsense transcripts 1 homolog
Length = 1180
Score = 30.8 bits (68), Expect = 1.6
Identities = 22/64 (34%), Positives = 32/64 (49%), Gaps = 10/64 (15%)
Query: 104 PDLINQIQPSDWQKAY------WEAHLQNCLDEAAEIALISPFKGRLADINISGTMLSCF 157
P+ I I P + Q+AY ++ L + L + EIA + F+GR DI I +SC
Sbjct: 767 PEQIGIITPYEGQRAYLVQYMQYQGSLHSRLYQEIEIASVDAFQGREKDIII----MSCV 822
Query: 158 LCNE 161
NE
Sbjct: 823 RSNE 826
>RNT1_SCHPO (Q09820) Regulator of nonsense transcripts 1 homolog
Length = 925
Score = 30.0 bits (66), Expect = 2.7
Identities = 21/64 (32%), Positives = 32/64 (49%), Gaps = 10/64 (15%)
Query: 104 PDLINQIQPSDWQKAYWEAHLQN------CLDEAAEIALISPFKGRLADINISGTMLSCF 157
P+ I + P D Q++Y ++QN L +A E+A + F+GR D I LSC
Sbjct: 715 PEQIGIVTPYDGQRSYIVQYMQNNGSMQKDLYKAVEVASVDAFQGREKDFII----LSCV 770
Query: 158 LCNE 161
+E
Sbjct: 771 RSSE 774
>YTFM_ECOLI (P39320) Hypothetical protein ytfM precursor
Length = 577
Score = 29.6 bits (65), Expect = 3.6
Identities = 11/32 (34%), Positives = 18/32 (55%)
Query: 72 DFSRDDSTNNKRKRSRDWNLNTAWQKLFELRW 103
D + +S + SR W+L++ WQ+ LRW
Sbjct: 338 DLNDTESDSTTLVASRYWDLSSGWQRAINLRW 369
>GPI3_YEAST (P32363) Phosphatidylinositol
N-acetylglucosaminyltransferase GPI3 subunit (EC
2.4.1.198) (GlcNAc-PI synthesis protein)
Length = 461
Score = 29.3 bits (64), Expect = 4.7
Identities = 29/108 (26%), Positives = 51/108 (46%), Gaps = 15/108 (13%)
Query: 8 SSLITLCIDHLANQLLFGEDEVIAIIPVIYE---LPSHLLDDLISRLTPHALYHFHLHMP 64
++L ++ ++ L L D VI + E + + L D+IS + P+A+
Sbjct: 139 NNLTSIWVNKLLTFTLTNIDRVICVSNTCKENMIVRTELSPDIIS-VIPNAV-------- 189
Query: 65 FQDVNEEDFSRDDSTNNKRKRSRDWNLNTAWQKLFELRWPDLINQIQP 112
V+E+ RD + KRK+SRD + +LF + DL+ +I P
Sbjct: 190 ---VSEDFKPRDPTGGTKRKQSRDKIVIVVIGRLFPNKGSDLLTRIIP 234
>FX21_HUMAN (O94952) F-box only protein 21
Length = 621
Score = 29.3 bits (64), Expect = 4.7
Identities = 11/38 (28%), Positives = 20/38 (51%)
Query: 78 STNNKRKRSRDWNLNTAWQKLFELRWPDLINQIQPSDW 115
S+ +R R + W++ F +RWP L+ P+D+
Sbjct: 56 SSTCRRLRELCQSSGKVWKEQFRVRWPSLMKHYSPTDY 93
>CADN_XENLA (P20310) Neural-cadherin 1 precursor (N-cadherin 1)
Length = 905
Score = 29.3 bits (64), Expect = 4.7
Identities = 12/40 (30%), Positives = 20/40 (50%)
Query: 50 RLTPHALYHFHLHMPFQDVNEEDFSRDDSTNNKRKRSRDW 89
++T + H H F V E FS ++N +++ RDW
Sbjct: 123 KITLRPPHEHHSHQGFHKVREIKFSTQRNSNGLQRQKRDW 162
>KAR9_YEAST (P32526) Karyogamy protein KAR9 (Cortical protein KAR9)
Length = 644
Score = 28.9 bits (63), Expect = 6.1
Identities = 30/124 (24%), Positives = 55/124 (44%), Gaps = 19/124 (15%)
Query: 15 IDHLANQLLFGEDEVIAIIPVIYELPSHLLDDLISRLTPHALYHFHLHMPFQDVNEEDFS 74
I L N+ G+D+++ + + L ++L IS + H + H+ D+ E++FS
Sbjct: 65 IPDLVNKKRLGKDDILLFMDWLL-LKKYMLYQFISDV--HNIEEGFAHL--LDLLEDEFS 119
Query: 75 RDDSTNNKRKRSRDWNLNTAWQKLFELRWPDLINQIQPSDWQKAYWEAHLQNCLDEAAEI 134
+DD ++K R + +F D+I + Q W +L+ LD + E
Sbjct: 120 KDDQDSDKYNR---------FSPMF-----DVIEESTQIKTQLEPWLTNLKELLDTSLEF 165
Query: 135 ALIS 138
IS
Sbjct: 166 NEIS 169
>FX21_MOUSE (Q8VDH1) F-box only protein 21
Length = 627
Score = 28.9 bits (63), Expect = 6.1
Identities = 11/38 (28%), Positives = 20/38 (51%)
Query: 78 STNNKRKRSRDWNLNTAWQKLFELRWPDLINQIQPSDW 115
S+ +R R + W++ F +RWP L+ P+D+
Sbjct: 56 SSTCRRLREVCQSSGQVWKEQFRVRWPSLMKHYSPTDY 93
>XYNA_RUMFL (P29126) Bifunctional endo-1,4-beta-xylanase xylA
precursor (EC 3.2.1.8)
Length = 954
Score = 28.5 bits (62), Expect = 8.0
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 7/52 (13%)
Query: 66 QDVNEEDFSRDDSTNNKRKRSRDWNLNTAWQKLFELRWPDLINQIQPSDWQK 117
Q N D++ ++ NN ++++ DWN W + W + NQ Q +DW +
Sbjct: 401 QQQNNWDWNNQNNWNNNQQQNNDWN---QWNN--QNNWNN--NQQQNNDWNQ 445
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,794,617
Number of Sequences: 164201
Number of extensions: 896114
Number of successful extensions: 1963
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1953
Number of HSP's gapped (non-prelim): 13
length of query: 173
length of database: 59,974,054
effective HSP length: 103
effective length of query: 70
effective length of database: 43,061,351
effective search space: 3014294570
effective search space used: 3014294570
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146681.8


