

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.2 - phase: 0
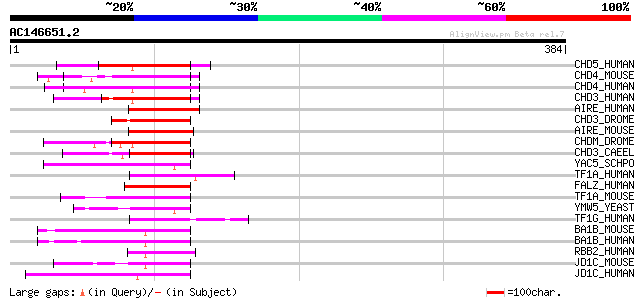
(384 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5 ... 82 2e-15
CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4 ... 79 2e-14
CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4 ... 79 3e-14
CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3 ... 75 2e-13
AIRE_HUMAN (O43918) Autoimmune regulator (Autoimmune polyendocri... 74 5e-13
CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3 72 3e-12
AIRE_MOUSE (Q9Z0E3) Autoimmune regulator (Autoimmune polyendocri... 72 3e-12
CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi... 70 7e-12
CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3 ... 64 6e-10
YAC5_SCHPO (Q09819) Hypothetical protein C16C9.05 in chromosome I 61 5e-09
TF1A_HUMAN (O15164) Transcription intermediary factor 1-alpha (T... 60 1e-08
FALZ_HUMAN (Q12830) Fetal Alzheimer antigen (Fetal Alz-50-reacti... 60 1e-08
TF1A_MOUSE (Q64127) Transcription intermediary factor 1-alpha (T... 59 2e-08
YMW5_YEAST (Q04779) Hypothetical 78.8 kDa protein in ABF2-CHL12 ... 59 2e-08
TF1G_HUMAN (Q9UPN9) Transcription intermediary factor 1-gamma (T... 58 4e-08
BA1B_MOUSE (Q9Z277) Bromodomain adjacent to zinc finger domain p... 58 4e-08
BA1B_HUMAN (Q9UIG0) Bromodomain adjacent to zinc finger domain p... 58 4e-08
RBB2_HUMAN (P29375) Retinoblastoma-binding protein 2 (RBBP-2) 58 5e-08
JD1C_MOUSE (P41230) Jumonji/ARID domain-containing protein 1C (S... 58 5e-08
JD1C_HUMAN (P41229) Jumonji/ARID domain-containing protein 1C (S... 58 5e-08
>CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5
(CHD-5)
Length = 1954
Score = 82.4 bits (202), Expect = 2e-15
Identities = 42/113 (37%), Positives = 60/113 (52%), Gaps = 6/113 (5%)
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE------CVI 86
SS +E + D AS S S ++ + + + KK+ DG YE C +
Sbjct: 289 SSSEEDEREESDFDSASIHSASVRSECSAALGKKSKRRRKKKRIDDGDGYETDHQDYCEV 348
Query: 87 CDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLD 139
C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P ++ D
Sbjct: 349 CQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEPKDDDD 401
Score = 72.4 bits (176), Expect = 2e-12
Identities = 28/64 (43%), Positives = 39/64 (60%)
Query: 62 KTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQ 121
K ++ + ++ D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W
Sbjct: 397 KDDDDEEEEGGCEEEEDDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWL 456
Query: 122 CPSC 125
CP C
Sbjct: 457 CPRC 460
>CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4
(CHD-4)
Length = 1915
Score = 79.0 bits (193), Expect = 2e-14
Identities = 45/121 (37%), Positives = 58/121 (47%), Gaps = 13/121 (10%)
Query: 20 KRKRRK-----LPIGPDQSSGKEQSNGKEDNSVASESSRSA----SAKRMLKTEEGTAQF 70
KRKR L + D S D S S SSRS +AK+ K EE
Sbjct: 297 KRKRSSSEDDDLDVESDFDDASINSYSVSDGST-SRSSRSRKKLRTAKKKKKGEEEVTAV 355
Query: 71 SSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEEND 130
+ H Y C +C GG ++ CD+CPR YH+ CLDP +++ P GKW CP C +E
Sbjct: 356 DGYETDHQDY---CEVCQQGGEIILCDTCPRAYHMVCLDPDMEKAPEGKWSCPHCEKEGI 412
Query: 131 Q 131
Q
Sbjct: 413 Q 413
Score = 71.2 bits (173), Expect = 4e-12
Identities = 35/88 (39%), Positives = 45/88 (50%), Gaps = 14/88 (15%)
Query: 38 QSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCD 97
Q KEDNS E + E G ++ D + C +C GG LLCCD
Sbjct: 413 QWEAKEDNSEGEE----------ILEEVG----GDPEEEDDHHMEFCRVCKDGGELLCCD 458
Query: 98 SCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+CP +YH+ CL+PPL IP G+W CP C
Sbjct: 459 TCPSSYHIHCLNPPLPEIPNGEWLCPRC 486
>CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4
(CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta)
Length = 1912
Score = 78.6 bits (192), Expect = 3e-14
Identities = 40/126 (31%), Positives = 64/126 (50%), Gaps = 19/126 (15%)
Query: 25 KLPIGPDQSSGKEQSNGKEDNSVASE---------------SSRSASAKRMLKTEEGTAQ 69
K+ +G S K S+ +D V S+ +SRS+ +++ L+T + +
Sbjct: 295 KIKLGGFGSKRKRSSSEDDDLDVESDFDDASINSYSVSDGSTSRSSRSRKKLRTTKKKKK 354
Query: 70 FSSKKKGHDGYFYE----CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+ DGY + C +C GG ++ CD+CPR YH+ CLDP +++ P GKW CP C
Sbjct: 355 GEEEVTAVDGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMEKAPEGKWSCPHC 414
Query: 126 FEENDQ 131
+E Q
Sbjct: 415 EKEGIQ 420
Score = 71.2 bits (173), Expect = 4e-12
Identities = 35/88 (39%), Positives = 44/88 (49%), Gaps = 14/88 (15%)
Query: 38 QSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCD 97
Q KEDNS E L+ E+ D + C +C GG LLCCD
Sbjct: 420 QWEAKEDNSEGEEILEEVGGD--LEEED------------DHHMEFCRVCKDGGELLCCD 465
Query: 98 SCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+CP +YH+ CL+PPL IP G+W CP C
Sbjct: 466 TCPSSYHIHCLNPPLPEIPNGEWLCPRC 493
>CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3
(CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha)
Length = 1944
Score = 75.5 bits (184), Expect = 2e-13
Identities = 40/106 (37%), Positives = 55/106 (51%), Gaps = 6/106 (5%)
Query: 31 DQSSGKEQS-NGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE----CV 85
D SG S +G+ D V ++ + R K G + +++ DGY + C
Sbjct: 325 DLDSGSVHSASGRPDGPVRTKKLKRGRPGRKKKKVLGCPAVAGEEEV-DGYETDHQDYCE 383
Query: 86 ICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQ 131
+C GG ++ CD+CPR YHL CLDP L R P GKW CP C +E Q
Sbjct: 384 VCQQGGEIILCDTCPRAYHLVCLDPELDRAPEGKWSCPHCEKEGVQ 429
Score = 70.1 bits (170), Expect = 9e-12
Identities = 29/62 (46%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Query: 64 EEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCP 123
EEG + +K+ D + C +C GG LLCCD+C +YH+ CL+PPL IP G+W CP
Sbjct: 441 EEGEEE--GEKEEEDDHMEYCRVCKDGGELLCCDACISSYHIHCLNPPLPDIPNGEWLCP 498
Query: 124 SC 125
C
Sbjct: 499 RC 500
>AIRE_HUMAN (O43918) Autoimmune regulator (Autoimmune
polyendocrinopathy candidiasis ectodermal dystrophy
protein) (APECED protein)
Length = 545
Score = 74.3 bits (181), Expect = 5e-13
Identities = 27/49 (55%), Positives = 33/49 (67%)
Query: 83 ECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQ 131
EC +C GG L+CCD CPR +HL CL PPL+ IP G W+C SC + Q
Sbjct: 298 ECAVCRDGGELICCDGCPRAFHLACLSPPLREIPSGTWRCSSCLQATVQ 346
>CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3
Length = 892
Score = 71.6 bits (174), Expect = 3e-12
Identities = 30/55 (54%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Query: 71 SSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+ K+K D + C +C GG+LLCCDSCP YH CL PPLK IP G W CP C
Sbjct: 27 AKKQKFRDEEY--CKVCSDGGDLLCCDSCPSVYHRTCLSPPLKSIPKGDWICPRC 79
>AIRE_MOUSE (Q9Z0E3) Autoimmune regulator (Autoimmune
polyendocrinopathy candidiasis ectodermal dystrophy
protein homolog) (APECED protein homolog)
Length = 552
Score = 71.6 bits (174), Expect = 3e-12
Identities = 25/45 (55%), Positives = 31/45 (68%)
Query: 83 ECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFE 127
EC +C GG L+CCD CPR +HL CL PPL+ IP G W+C C +
Sbjct: 300 ECAVCHDGGELICCDGCPRAFHLACLSPPLQEIPSGLWRCSCCLQ 344
Score = 30.8 bits (68), Expect = 6.0
Identities = 13/42 (30%), Positives = 18/42 (41%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C +C G +L C C +H +C P P +C SC
Sbjct: 434 CSVCGDGTEVLRCAHCAAAFHWRCHFPTAAARPGTNLRCKSC 475
>CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi-2
homolog (dMi-2)
Length = 1982
Score = 70.5 bits (171), Expect = 7e-12
Identities = 42/118 (35%), Positives = 55/118 (46%), Gaps = 17/118 (14%)
Query: 24 RKLPIGPDQSSGKEQS-NGKEDNSVASESSRSASA---KRMLKTEEGTAQFSSKKK---- 75
R L D + KE + K DNS + + A ++ KT+ G +F K K
Sbjct: 305 RMLQKSDDSADEKEAPVSSKADNSAPAAQDDGSGAPVVRKKAKTKIGN-KFKKKNKLKKT 363
Query: 76 -----GHDGYFYE---CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
G DG C +C GG ++ CD+CPR YHL CL+P L P GKW CP C
Sbjct: 364 KNFPEGEDGEHEHQDYCEVCQQGGEIILCDTCPRAYHLVCLEPELDEPPEGKWSCPHC 421
Score = 68.9 bits (167), Expect = 2e-11
Identities = 27/55 (49%), Positives = 36/55 (65%)
Query: 71 SSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+++++ D + C +C GG LLCCDSCP YH CL+PPL IP G W+CP C
Sbjct: 427 AAEEEDDDEHQEFCRVCKDGGELLCCDSCPSAYHTFCLNPPLDTIPDGDWRCPRC 481
>CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3
homolog (CHD-3)
Length = 1787
Score = 63.9 bits (154), Expect = 6e-10
Identities = 23/42 (54%), Positives = 28/42 (65%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C IC N+L CD+CP +YH C+DPPL IP G+W CP C
Sbjct: 331 CRICKETSNILLCDTCPSSYHAYCIDPPLTEIPEGEWSCPRC 372
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/93 (29%), Positives = 50/93 (53%), Gaps = 3/93 (3%)
Query: 37 EQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKG--HDGYFYECVICDLGGNLL 94
E GKE+ + +++ K L++ + + + K++G + + C +C+ G L+
Sbjct: 220 EAEKGKEEARINRAAAKVDKRKAALESARASKR-ARKEQGVVEENHQENCEVCNQDGELM 278
Query: 95 CCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFE 127
CD+C R YH+ C+D +++ P G W CP C E
Sbjct: 279 LCDTCTRAYHVACIDENMEQPPEGDWSCPHCEE 311
>YAC5_SCHPO (Q09819) Hypothetical protein C16C9.05 in chromosome I
Length = 404
Score = 60.8 bits (146), Expect = 5e-09
Identities = 33/105 (31%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Query: 24 RKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTE-EGTAQFSSKKKGHDGYFY 82
+++ I K + K N + SR +S K L+TE + T ++K
Sbjct: 59 KEMDISSPVKKQKASYSNKSPNKAPIQKSRGSSLKSHLETESQQTPVKRRRRKATIRNVD 118
Query: 83 ECVICDLGGNLLCCDSCPRTYHLQCLDPPL--KRIPMGKWQCPSC 125
C C G +CC+ CP ++HL CL+PPL + IP G W C +C
Sbjct: 119 YCSACGGRGLFICCEGCPCSFHLSCLEPPLTPENIPEGSWFCVTC 163
>TF1A_HUMAN (O15164) Transcription intermediary factor 1-alpha
(TIF1-alpha) (Tripartite motif-containing protein 24)
Length = 1050
Score = 59.7 bits (143), Expect = 1e-08
Identities = 26/78 (33%), Positives = 36/78 (45%), Gaps = 6/78 (7%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFE------ENDQLKPLNN 137
C +C GG LLCC+ CP+ +HL C P L P G+W C C + E D P +N
Sbjct: 829 CAVCQNGGELLCCEKCPKVFHLSCHVPTLTNFPSGEWICTFCRDLSKPEVEYDCDAPSHN 888
Query: 138 LDSISRRARTKTVPVKSK 155
+ K P+ +
Sbjct: 889 SEKKKTEGLVKLTPIDKR 906
>FALZ_HUMAN (Q12830) Fetal Alzheimer antigen (Fetal Alz-50-reactive
clone 1)
Length = 810
Score = 59.7 bits (143), Expect = 1e-08
Identities = 21/46 (45%), Positives = 30/46 (64%)
Query: 80 YFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
Y C +C G+LLCC++C YHL+C+ PPL+ +P +WQC C
Sbjct: 250 YDDHCRVCHKLGDLLCCETCSAVYHLECVKPPLEEVPEDEWQCEVC 295
>TF1A_MOUSE (Q64127) Transcription intermediary factor 1-alpha
(TIF1-alpha) (Tripartite motif-containing protein 24)
Length = 1051
Score = 59.3 bits (142), Expect = 2e-08
Identities = 30/90 (33%), Positives = 39/90 (43%), Gaps = 14/90 (15%)
Query: 36 KEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLC 95
+ SNGK + S AS+ S +K D C +C GG LLC
Sbjct: 796 ENSSNGKSEWSDASQKS--------------PVHVGETRKEDDPNEDWCAVCQNGGELLC 841
Query: 96 CDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
C+ CP+ +HL C P L P G+W C C
Sbjct: 842 CEKCPKVFHLTCHVPTLTNFPSGEWICTFC 871
>YMW5_YEAST (Q04779) Hypothetical 78.8 kDa protein in ABF2-CHL12
intergenic region
Length = 684
Score = 58.9 bits (141), Expect = 2e-08
Identities = 29/83 (34%), Positives = 41/83 (48%), Gaps = 12/83 (14%)
Query: 45 NSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYH 104
NS+ S S+ AS K T F ++ C C+ G+ LCCD+CP+++H
Sbjct: 234 NSIKSTSN--ASEKIFRDKNNSTIDFENEDF--------CSACNQSGSFLCCDTCPKSFH 283
Query: 105 LQCLDPPL--KRIPMGKWQCPSC 125
CLDPP+ +P G W C C
Sbjct: 284 FLCLDPPIDPNNLPKGDWHCNEC 306
>TF1G_HUMAN (Q9UPN9) Transcription intermediary factor 1-gamma
(TIF1-gamma) (RET-fused gene 7 protein) (Rfg7 protein)
(Tripartite motif-containing protein 33)
Length = 1127
Score = 58.2 bits (139), Expect = 4e-08
Identities = 30/82 (36%), Positives = 41/82 (49%), Gaps = 6/82 (7%)
Query: 84 CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLDSISR 143
C +C GG+LLCC+ CP+ +HL C P L P G W C C D KP D +
Sbjct: 890 CAVCQNGGDLLCCEKCPKVFHLTCHVPTLLSFPSGDWICTFC---RDIGKPEVEYDCDNL 946
Query: 144 RARTKTVPVKSKAGVNPVNLEK 165
+ K K+ G++PV+ K
Sbjct: 947 QHSKKG---KTAQGLSPVDQRK 965
>BA1B_MOUSE (Q9Z277) Bromodomain adjacent to zinc finger domain
protein 1B (Williams-Beuren syndrome chromosome region 9
protein homolog) (WBRS9)
Length = 1479
Score = 58.2 bits (139), Expect = 4e-08
Identities = 37/121 (30%), Positives = 53/121 (43%), Gaps = 20/121 (16%)
Query: 20 KRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDG 79
K+K+RKL QS +S ++ E ++ ASA KT AQ S+ G
Sbjct: 1116 KQKKRKL-----QSEDSTKSEEVDEEKKMVEEAKVASALEKWKTAIREAQTFSRMHVLLG 1170
Query: 80 YFYECVICDLGGN---------------LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPS 124
C+ D+ L+ CD C + +HL CL P L +P G+WQCP+
Sbjct: 1171 MLDACIKWDMSAENARCKVCRKKGEDDKLILCDECNKAFHLFCLRPALYEVPDGEWQCPA 1230
Query: 125 C 125
C
Sbjct: 1231 C 1231
>BA1B_HUMAN (Q9UIG0) Bromodomain adjacent to zinc finger domain
protein 1B (Williams-Beuren syndrome chromosome region 9
protein) (WBRS9) (Williams syndrome transcription factor)
(hWALP2)
Length = 1483
Score = 58.2 bits (139), Expect = 4e-08
Identities = 39/121 (32%), Positives = 54/121 (44%), Gaps = 20/121 (16%)
Query: 20 KRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDG 79
K+KRRKL + S K + +E V E ++ ASA KT AQ S+ G
Sbjct: 1116 KQKRRKLQ---SEDSAKTEEVDEEKKMV--EEAKVASALEKWKTAIREAQTFSRMHVLLG 1170
Query: 80 YFYECVICDLGGN---------------LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPS 124
C+ D+ L+ CD C + +HL CL P L +P G+WQCP+
Sbjct: 1171 MLDACIKWDMSAENARCKVCRKKGEDDKLILCDECNKAFHLFCLRPALYEVPDGEWQCPA 1230
Query: 125 C 125
C
Sbjct: 1231 C 1231
>RBB2_HUMAN (P29375) Retinoblastoma-binding protein 2 (RBBP-2)
Length = 1722
Score = 57.8 bits (138), Expect = 5e-08
Identities = 24/50 (48%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query: 82 YECVICDLGGN---LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEE 128
Y C+ C G N LL CD C +YH CL PPL +P G W+CP C E
Sbjct: 294 YVCMFCGRGNNEDKLLLCDGCDDSYHTFCLIPPLPDVPKGDWRCPKCVAE 343
>JD1C_MOUSE (P41230) Jumonji/ARID domain-containing protein 1C (SmcX
protein) (Xe169 protein)
Length = 1554
Score = 57.8 bits (138), Expect = 5e-08
Identities = 34/98 (34%), Positives = 44/98 (44%), Gaps = 13/98 (13%)
Query: 31 DQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLG 90
+ +S K GKE+ S + E + + L+ AQF Y C +C G
Sbjct: 284 ESTSPKTFLEGKEELSHSPEPCTKMTMR--LRRNHSNAQFIES--------YVCRMCSRG 333
Query: 91 GN---LLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
LL CD C YH+ CL PPL IP G W+CP C
Sbjct: 334 DEDDKLLLCDGCDDNYHIFCLLPPLPEIPKGVWRCPKC 371
>JD1C_HUMAN (P41229) Jumonji/ARID domain-containing protein 1C (SmcX
protein) (Xe169 protein)
Length = 1560
Score = 57.8 bits (138), Expect = 5e-08
Identities = 35/122 (28%), Positives = 56/122 (45%), Gaps = 8/122 (6%)
Query: 12 MLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFS 71
++ K+ L++K ++ P P KE+ G S + S + + + E + +
Sbjct: 250 LMAKDKTLRKKDKEGPECPPTVVVKEELGGDVKVESTSPKTFLESKEELSHSPEPCTKMT 309
Query: 72 SK-KKGH-DGYFYECVIC------DLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCP 123
+ ++ H + F E +C D LL CD C YH+ CL PPL IP G W+CP
Sbjct: 310 MRLRRNHSNAQFIESYVCRMCSRGDEDDKLLLCDGCDDNYHIFCLLPPLPEIPKGVWRCP 369
Query: 124 SC 125
C
Sbjct: 370 KC 371
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,485,529
Number of Sequences: 164201
Number of extensions: 1985085
Number of successful extensions: 5189
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 4964
Number of HSP's gapped (non-prelim): 267
length of query: 384
length of database: 59,974,054
effective HSP length: 112
effective length of query: 272
effective length of database: 41,583,542
effective search space: 11310723424
effective search space used: 11310723424
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146651.2


