

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146651.1 - phase: 0
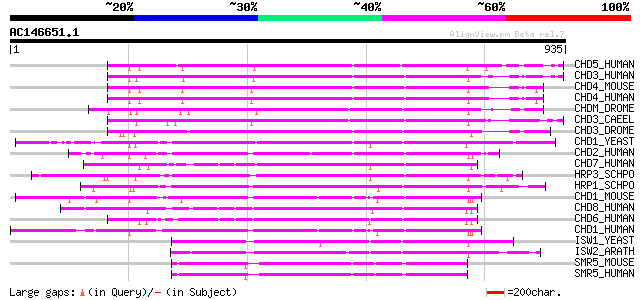
(935 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5 ... 538 e-152
CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3 ... 536 e-151
CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4 ... 530 e-150
CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4 ... 530 e-150
CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi... 501 e-141
CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3 ... 495 e-139
CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3 471 e-132
CHD1_YEAST (P32657) Chromo domain protein 1 464 e-130
CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2 ... 464 e-130
CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7 ... 462 e-129
HRP3_SCHPO (O14139) Chromodomain helicase hrp3 453 e-127
HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1 453 e-126
CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1 ... 436 e-121
CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8 ... 432 e-120
CHD6_HUMAN (Q8TD26) Chromodomain-helicase-DNA-binding protein 6 ... 431 e-120
CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1 ... 431 e-120
ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain I... 375 e-103
ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPas... 370 e-102
SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin depe... 357 8e-98
SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin depe... 357 8e-98
>CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5
(CHD-5)
Length = 1954
Score = 538 bits (1385), Expect = e-152
Identities = 351/845 (41%), Positives = 474/845 (55%), Gaps = 105/845 (12%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
EF VKW G S+ H SW+ E L++ NY+ K YG+ + E+ K
Sbjct: 510 EFFVKWAGLSYWHCSWVKELQLELYHTVMYRNYQRKNDMDEPPPFDYGSGDEDGKSEKRK 569
Query: 215 NPE-----------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWE--SL 249
N + R+L K+G +KW PY++CTWE +
Sbjct: 570 NKDPLYAKMEERFYRYGIKPEWMMIHRILNHSFDKKGDVHYLIKWKDLPYDQCTWEIDDI 629
Query: 250 DEPVLQNSSHLI---TRFNMFETLTLEREASKENSTKKSSDRQ---------NDIVNLLE 297
D P N + E L + K+ K D+Q + V +
Sbjct: 630 DIPYYDNLKQAYWGHRELMLGEDTRLPKRLLKKGK-KLRDDKQEKPPDTPIVDPTVKFDK 688
Query: 298 QP--KELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKV 355
QP + GG+L P+QLE LNWLR W + + ILADEMGLGKT+ F+ SLY E
Sbjct: 689 QPWYIDSTGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHS 748
Query: 356 SRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD---PSGLN- 411
P LV PL T+ NW EF +WAPD VV Y G ++R++IR+ E+ D SG
Sbjct: 749 KGPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGKKV 808
Query: 412 ---KKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISF 468
KK KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LNS
Sbjct: 809 FRMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSYKI 868
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHM 528
+++LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHM
Sbjct: 869 DYKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHM 928
Query: 529 LRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVM 588
LRRLK D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M
Sbjct: 929 LRRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGG-NQVSLLNIMM 987
Query: 589 QLRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFS 645
L+K CNHPYL P P + + +K+S KL LL MLK L EGHRVLIFS
Sbjct: 988 DLKKCCNHPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFS 1047
Query: 646 QMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGL 704
QMTK+LD+LED+L E+ YER+DG ++ RQ AI RFN +F FLLSTR+ GL
Sbjct: 1048 QMTKMLDLLEDFL--EYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGL 1105
Query: 705 GINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKK 764
GINLATADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AK+K
Sbjct: 1106 GINLATADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRK 1165
Query: 765 LMLDQL-----FKGKSGS--QKEVEDILKWGTEELFND---SCALNGK----DTSENNNS 810
+ML L KSGS ++E++DILK+GTEELF D G+ + +S
Sbjct: 1166 MMLTHLVVRPGLGSKSGSMTKQELDDILKFGTEELFKDDVEGMMSQGQRPVTPIPDVQSS 1225
Query: 811 NKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAE 870
A + KH G ++K ++SS I +D+ AI KLLDR+ QDA TD E
Sbjct: 1226 KGGNLAASAKKKHGSTPPG-----DNKDVEDSSVIHYDDAAISKLLDRN--QDA-TDDTE 1277
Query: 871 GDSENDMLGSMKALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEW 928
+ N+ L S K ++ +E E VE E K+E+N + + W
Sbjct: 1278 LQNMNEYLSSFKVAQYVVREEDGVEEVERE-------------IIKQEENV----DPDYW 1320
Query: 929 DRLLR 933
++LLR
Sbjct: 1321 EKLLR 1325
>CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3
(CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha)
Length = 1944
Score = 536 bits (1380), Expect = e-151
Identities = 343/834 (41%), Positives = 461/834 (55%), Gaps = 124/834 (14%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEE--- 211
EF VKWVG S+ H SW E L++ NY+ K YG+ + +
Sbjct: 549 EFFVKWVGLSYWHCSWAKELQLEIFHLVMYRNYQRKNDMDEPPPLDYGSGEDDGKSDKRK 608
Query: 212 --------------------QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDE 251
+W R++ K+G VKW PY++ TWE +
Sbjct: 609 VKDPHYAEMEEKYYRFGIKPEWMTVHRIINHSVDKKGNYHYLVKWRDLPYDQSTWEEDEM 668
Query: 252 PVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQND----------IVNLLEQPKE 301
+ + H + + E + E A KK + Q D V QP+
Sbjct: 669 NIPEYEEHKQSYWRHRELIMGEDPAQPRKYKKKKKELQGDGPPSSPTNDPTVKYETQPRF 728
Query: 302 LR--GGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPC 359
+ GG+L +QLE LNWLR W + + ILADEMGLGKTI F+ SLY E P
Sbjct: 729 ITATGGTLHMYQLEGLNWLRFSWAQGTDTILADEMGLGKTIQTIVFLYSLYKEGHTKGPF 788
Query: 360 LVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPS------GLNKK 413
LV PL T+ NW EF +WAP VV Y G +RAIIR+ E+ D + K
Sbjct: 789 LVSAPLSTIINWEREFQMWAPKFYVVTYTGDKDSRAIIRENEFSFEDNAIKGGKKAFKMK 848
Query: 414 TEAY-KFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRV 472
EA KF+VLLTSYE++ D + + W L+VDE HRLKN++SK F +LN H++
Sbjct: 849 REAQVKFHVLLTSYELITIDQAALGSIRWACLVVDEAHRLKNNQSKFFRVLNGYKIDHKL 908
Query: 473 LLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRL 532
LLTGTPLQNNL E+++LLNFL P F +L F E F D++ +++ +L L+ PHMLRRL
Sbjct: 909 LLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADISKEDQIKKLHDLLGPHMLRRL 968
Query: 533 KKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRK 592
K D +N+P KTE +V VELS +Q +YY+ +LT+N++ L + G G Q S+LNI+M L+K
Sbjct: 969 KADVFKNMPAKTELIVRVELSPMQKKYYKYILTRNFEALNSRGGG-NQVSLLNIMMDLKK 1027
Query: 593 VCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTK 649
CNHPYL P P S + IK+S KL LL ML+ L ++GHRVLIFSQMTK
Sbjct: 1028 CCNHPYLFPVAAMESPKLPSGAYEGGALIKSSGKLMLLQKMLRKLKEQGHRVLIFSQMTK 1087
Query: 650 LLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINL 708
+LD+LED+L+ E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINL
Sbjct: 1088 MLDLLEDFLDYE-GYK-YERIDGGITGALRQEAIDRFNAPGAQQFCFLLSTRAGGLGINL 1145
Query: 709 ATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLD 768
ATADTVII+DSD+NPH DIQA +RAHRIGQ+N++++YR V RASVEERI Q+AK+K+ML
Sbjct: 1146 ATADTVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERITQVAKRKMMLT 1205
Query: 769 QLF-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEH 821
L K S S++E++DILK+GTEELF D EN NK+E
Sbjct: 1206 HLVVRPGLGSKAGSMSKQELDDILKFGTEELFKD----------ENEGENKEE------- 1248
Query: 822 KHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSM 881
+SS I +D AI +LLDR+ QDA+ D + + N+ L S
Sbjct: 1249 -------------------DSSVIHYDNEAIARLLDRN--QDATED-TDVQNMNEYLSSF 1286
Query: 882 KALEW--NDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEENEWDRLLR 933
K ++ +E E +E E K+E+N + + W++LLR
Sbjct: 1287 KVAQYVVREEDKIEEIERE-------------IIKQEENV----DPDYWEKLLR 1323
>CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4
(CHD-4)
Length = 1915
Score = 530 bits (1366), Expect = e-150
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 535 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 594
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 595 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 654
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 655 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 713
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 714 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 773
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 774 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 833
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 834 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 893
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 894 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 953
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 954 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1012
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1013 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1072
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1073 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1130
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1131 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1190
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1191 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1234
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1235 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1275
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1276 AQYVVREEEMGEEEEVERE 1294
>CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4
(CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta)
Length = 1912
Score = 530 bits (1366), Expect = e-150
Identities = 337/799 (42%), Positives = 448/799 (55%), Gaps = 104/799 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAK----------YGTATINICEEQWK 214
+F VKW G S+ H SW+SE L++ + NY+ K +G + + K
Sbjct: 542 QFFVKWQGMSYWHCSWVSELQLELHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNK 601
Query: 215 NPE---------------------RLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPV 253
+P+ R+L K+G +KW PY++ +WES D +
Sbjct: 602 DPKFAEMEERFYRYGIKPEWMMIHRILNHSVDKKGHVHYLIKWRDLPYDQASWESEDVEI 661
Query: 254 LQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQ----------NDIVNLLEQPKEL- 302
+ +N E + E E KK R+ + V QP+ L
Sbjct: 662 QDYDLFKQSYWNHRELMRGE-EGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLD 720
Query: 303 -RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLV 361
GG+L P+Q+E LNWLR W + + ILADEMGLGKT+ F+ SLY E P LV
Sbjct: 721 ATGGTLHPYQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLV 780
Query: 362 LVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD-------PSGLNKKT 414
PL T+ NW EF +WAPD+ VV Y G +RAIIR+ E+ D + KK
Sbjct: 781 SAPLSTIINWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKE 840
Query: 415 EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLL 474
+ KF+VLLTSYE++ D + + W LIVDE HRLKN++SK F +LN S QH++LL
Sbjct: 841 ASVKFHVLLTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLL 900
Query: 475 TGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKK 534
TGTPLQNNL E+++LLNFL P F +L F E F D+ +++ +L ++ PHMLRRLK
Sbjct: 901 TGTPLQNNLEELFHLLNFLTPERFHNLEGFLEEFADIAKEDQIKKLHDMLGPHMLRRLKA 960
Query: 535 DAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVC 594
D +N+P KTE +V VELS +Q +YY+ +LT+N++ L G G Q S+LN+VM L+K C
Sbjct: 961 DVFKNMPSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGG-NQVSLLNVVMDLKKCC 1019
Query: 595 NHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLL 651
NHPYL P P + + I+AS KL LL MLK L + GHRVLIFSQMTK+L
Sbjct: 1020 NHPYLFPVAAMEAPKMPNGMYDGSALIRASGKLLLLQKMLKNLKEGGHRVLIFSQMTKML 1079
Query: 652 DILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLAT 710
D+LED+L E G K YER+DG ++ RQ AI RFN +F FLLSTR+ GLGINLAT
Sbjct: 1080 DLLEDFLEHE-GYK-YERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLAT 1137
Query: 711 ADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQL 770
ADTVIIYDSD+NPH DIQA +RAHRIGQ+ ++++YR V RASVEERI Q+AKKK+ML L
Sbjct: 1138 ADTVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHL 1197
Query: 771 F-------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKH 823
K S S++E++DILK+GTEELF D G D E
Sbjct: 1198 VVRPGLGSKTGSMSKQELDDILKFGTEELFKDEATDGGGDNKEG---------------- 1241
Query: 824 RKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKA 883
++SS I +D+ AI +LLDR+ QD + D E N+ L S K
Sbjct: 1242 ----------------EDSSVIHYDDKAIERLLDRN--QDETED-TELQGMNEYLSSFKV 1282
Query: 884 LEW----NDEPTEEHVEGE 898
++ + EE VE E
Sbjct: 1283 AQYVVREEEMGEEEEVERE 1301
>CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi-2
homolog (dMi-2)
Length = 1982
Score = 501 bits (1290), Expect = e-141
Identities = 323/853 (37%), Positives = 456/853 (52%), Gaps = 138/853 (16%)
Query: 133 LETKQKEVVLEKGMGSSGDNKVQDSIGEEVSY----EFLVKWVGKSHIHNSWISESHLKV 188
L K ++++ + S D+ S G + S E+ +KW S+ H W+ E L V
Sbjct: 486 LTGKAEKIITWRWAQRSNDDGPSTSKGSKNSNSRVREYFIKWHNMSYWHCEWVPEVQLDV 545
Query: 189 IAKRKLENYKAKY--------------------------------GTATINICEE----- 211
+ +++ KY + EE
Sbjct: 546 HHPLMIRSFQRKYDMEEPPKFEESLDEADTRYKRIQRHKDKVGMKANDDAEVLEERFYKN 605
Query: 212 ----QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMF 267
+W +R++ RT++ G++ VKW PY++ TWE + + + ++
Sbjct: 606 GVKPEWLIVQRVINHRTARDGSTMYLVKWRELPYDKSTWEEEGDDIQGLRQAIDYYQDLR 665
Query: 268 ETLTLEREASKENSTKKS--------SDRQNDIVNLLEQPK-----------------EL 302
T E S+ +KK D + + P+ E
Sbjct: 666 AVCTSETTQSRSKKSKKGRKSKLKVEDDEDRPVKHYTPPPEKPTTDLKKKYEDQPAFLEG 725
Query: 303 RGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVL 362
G L P+Q+E +NWLR W + + ILADEMGLGKTI F+ SLY E P LV
Sbjct: 726 TGMQLHPYQIEGINWLRYSWGQGIDTILADEMGLGKTIQTVTFLYSLYKEGHCRGPFLVA 785
Query: 363 VPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKK-----TEAY 417
VPL T+ NW EF LWAPD + Y G +RA+IR+ E + + K T Y
Sbjct: 786 VPLSTLVNWEREFELWAPDFYCITYIGDKDSRAVIRENELSFEEGAIRGSKVSRLRTTQY 845
Query: 418 KFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGT 477
KFNVLLTSYE++ D + + W VL+VDE HRLK+++SK F +LNS + +++LLTGT
Sbjct: 846 KFNVLLTSYELISMDAACLGSIDWAVLVVDEAHRLKSNQSKFFRILNSYTIAYKLLLTGT 905
Query: 478 PLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAM 537
PLQNNL E+++LLNFL F L AF+ F D++ E+V L +++ PHMLRRLK D +
Sbjct: 906 PLQNNLEELFHLLNFLSRDKFNDLQAFQGEFADVSKEEQVKRLHEMLGPHMLRRLKTDVL 965
Query: 538 QNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHP 597
+N+P K+E +V VELS++Q ++Y+ +LTKNY+ L N G S++NI+M L+K CNHP
Sbjct: 966 KNMPSKSEFIVRVELSAMQKKFYKFILTKNYEAL-NSKSGGGSCSLINIMMDLKKCCNHP 1024
Query: 598 YLIP-GTEPDSGSVEFLHEMR--IKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDIL 654
YL P E + + L+E+ KA+ KL LL MLK L + HRVLIFSQMTK+LDIL
Sbjct: 1025 YLFPSAAEEATTAAGGLYEINSLTKAAGKLVLLSKMLKQLKAQNHRVLIFSQMTKMLDIL 1084
Query: 655 EDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN-QDKSRFVFLLSTRSCGLGINLATADT 713
ED+L E YER+DG ++ T RQ AI RFN +FVFLLSTR+ GLGINLATADT
Sbjct: 1085 EDFLEGE--QYKYERIDGGITGTLRQEAIDRFNAPGAQQFVFLLSTRAGGLGINLATADT 1142
Query: 714 VIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLF-- 771
VIIYDSD+NPH DIQA +RAHRIGQ+N++++YR V R SVEER+ Q+AK+K+ML L
Sbjct: 1143 VIIYDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRNSVEERVTQVAKRKMMLTHLVVR 1202
Query: 772 -----KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKR 826
KG + +++E++DIL++GTE+LF + +K+EA
Sbjct: 1203 PGMGGKGANFTKQELDDILRFGTEDLFKE--------------DDKEEA----------- 1237
Query: 827 TGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDS-ENDMLGSMKALE 885
I +D+ A+ +LLDR+N I E +S N+ L S K
Sbjct: 1238 ------------------IHYDDKAVAELLDRTN-----RGIEEKESWANEYLSSFKVAS 1274
Query: 886 WNDEPTEEHVEGE 898
+ + EE E E
Sbjct: 1275 YATKEEEEEEETE 1287
>CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3
homolog (CHD-3)
Length = 1787
Score = 495 bits (1274), Expect = e-139
Identities = 317/840 (37%), Positives = 451/840 (52%), Gaps = 117/840 (13%)
Query: 165 EFLVKWVGKSHIHNSWISESHLKVIAKRKLENYKAKYGTATINICEE------------- 211
EF VKW ++ W+SE+ + V + Y K + I EE
Sbjct: 426 EFFVKWKYLAYWQCEWLSETLMDVYFTALVRMYWRKVDSENPPIFEESTLSRHHSDHDPY 485
Query: 212 -------------QWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSS 258
+W R++ + + + VKW Y TWE D +
Sbjct: 486 KLRERFYQYGVKPEWMQIHRIINHLSYAKSQQDYLVKWKELSYEHATWERDDTDIANYED 545
Query: 259 HLITRF-------------NMFETLTLEREAS------KENSTKKSSDRQNDIVNLLE-Q 298
+I + N+ + + +REA E ++++ + DI+ E Q
Sbjct: 546 AIIKYWHHRERMLNDEVPRNVQKMIAKQREAKGLGPKEDEVTSRRKKREKIDILKKYEVQ 605
Query: 299 PKELR--GGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVS 356
P + GG+L P+QLE +NWLR CW + ILADEMGLGKT+ + F+ +L E
Sbjct: 606 PDFISETGGNLHPYQLEGINWLRHCWSNGTDAILADEMGLGKTVQSLTFLYTLMKEGHTK 665
Query: 357 RPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD------PSGL 410
P L+ PL T+ NW E LW PD VV Y G ++R +IR++E+ D P
Sbjct: 666 GPFLIAAPLSTIINWEREAELWCPDFYVVTYVGDRESRMVIREHEFSFVDGAVRGGPKVS 725
Query: 411 NKKT-EAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQ 469
KT E KF+VLLTSYE + D + + W L+VDE HRLKN++S F L + Q
Sbjct: 726 KIKTLENLKFHVLLTSYECINMDKAILSSIDWAALVVDEAHRLKNNQSTFFKNLREYNIQ 785
Query: 470 HRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHML 529
+RVLLTGTPLQNNL E+++LLNFL P F L +F F++++ +++++L L+ PHML
Sbjct: 786 YRVLLTGTPLQNNLEELFHLLNFLAPDRFNQLESFTAEFSEISKEDQIEKLHNLLGPHML 845
Query: 530 RRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQ 589
RRLK D + +P K E +V VELS++Q +YY+ +LT+N+ L N+ G Q S++NI+M+
Sbjct: 846 RRLKADVLTGMPSKQELIVRVELSAMQKKYYKNILTRNFDAL-NVKNGGTQMSLINIIME 904
Query: 590 LRKVCNHPYLIPGT---EPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQ 646
L+K CNHPYL P + + IK + K LL ML+ L GHRVLIFSQ
Sbjct: 905 LKKCCNHPYLFMKACLEAPKLKNGMYEGSALIKNAGKFVLLQKMLRKLKDGGHRVLIFSQ 964
Query: 647 MTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSR-FVFLLSTRSCGLG 705
MT +LDILED+ ++E G K YER+DGS++ RQ AI R+N ++ FVFLLSTR+ GLG
Sbjct: 965 MTMMLDILEDFCDVE-GYK-YERIDGSITGQQRQDAIDRYNAPGAKQFVFLLSTRAGGLG 1022
Query: 706 INLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKL 765
INLATADTVIIYDSD+NPH DIQA +RAHR+GQ +++++YR V + SVEERI +AKKK+
Sbjct: 1023 INLATADTVIIYDSDWNPHNDIQAFSRAHRLGQKHKVMIYRFVTKGSVEERITSVAKKKM 1082
Query: 766 MLDQLF--------KGKSGSQKEVEDILKWGTEELFNDSCA-LNGKDTSENNNSNKDEAV 816
+L L GKS S+ E++D+L+WGTEELF + A + G D E +S K
Sbjct: 1083 LLTHLVVRAGLGAKDGKSMSKTELDDVLRWGTEELFKEEEAPVEGAD-GEGTSSKK---- 1137
Query: 817 AEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLLDRSNLQDASTDIAEGDSEND 876
N +I+WD+ A+ LLDR+ ++ + N+
Sbjct: 1138 -----------------------PNEQEIVWDDAAVDFLLDRNKEEEGQDGEKKEHWTNE 1174
Query: 877 MLGSMKALEWNDEPTEEHVEGESPPHGADDMCTQNSEKKEDNAVIGGEE---NEWDRLLR 933
L S K +N + ++ ADD + KE G EE N W++LL+
Sbjct: 1175 YLSSFKVATYNTKEADD----------ADDDEDETEVIKE-----GTEEQDPNYWEKLLK 1219
>CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3
Length = 892
Score = 471 bits (1211), Expect = e-132
Identities = 300/780 (38%), Positives = 420/780 (53%), Gaps = 90/780 (11%)
Query: 165 EFLVKWVGKSHIHNSWISES-----HLKVIA---------KRKLENYKAKYGTATINI-- 208
E+ +KW G S+ H WI E H ++A + LE + G
Sbjct: 112 EYFIKWHGMSYWHCEWIPEGQMLLHHASMVASFQRRSDMEEPSLEELDDQDGNLHERFYR 171
Query: 209 --CEEQWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNM 266
+ +W +R++ G + VKW YN+ +WE + + + I +
Sbjct: 172 YGIKPEWLLVQRVINHSEEPNGGTMYLVKWRELSYNDSSWERESDSI-PGLNQAIALYKK 230
Query: 267 FETLTLEREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSR 326
+ R+ + T + + D L++ G L P Q+E ++WLR W +
Sbjct: 231 LRSSNKGRQRDRPAPTIDLNKKYEDQPVFLKEA----GLKLHPFQIEGVSWLRYSWGQGI 286
Query: 327 NVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQ 386
ILADEMGLGKTI F+ SL+ E P L+ VPL T+ NW E LWAP++ V
Sbjct: 287 PTILADEMGLGKTIQTVVFLYSLFKEGHCRGPFLISVPLSTLTNWERELELWAPELYCVT 346
Query: 387 YHGCAKARAIIRQYE--WHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVL 444
Y G ARA+IR++E + + + YKFNV+LTSYE + D + + W L
Sbjct: 347 YVGGKTARAVIRKHEISFEEVTTKTMRENQTQYKFNVMLTSYEFISVDAAFLGCIDWAAL 406
Query: 445 IVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAF 504
+VDE HRL++++SK F +L+ +++LLTGTPLQNNL E+++LLNFL F L F
Sbjct: 407 VVDEAHRLRSNQSKFFRILSKYRIGYKLLLTGTPLQNNLEELFHLLNFLSSGKFNDLQTF 466
Query: 505 EERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAML 564
+ F D++ E+V L +++ PHMLRRLK D ++++PPK+E +V VELSS+Q ++Y+ +L
Sbjct: 467 QAEFTDVSKEEQVKRLHEILEPHMLRRLKADVLKSMPPKSEFIVRVELSSMQKKFYKHIL 526
Query: 565 TKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIP-GTEPDSGSVEFLHEMR--IKAS 621
TKN++ L G G S+LNI+M LRK CNHPYL P E + S L+EM KAS
Sbjct: 527 TKNFKALNQKGGG-RVCSLLNIMMDLRKCCNHPYLFPSAAEEATISPSGLYEMSSLTKAS 585
Query: 622 AKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQT 681
KL LL MLK L + HRVL+FSQMTK+L++LE +L E Y+R+DGS+ RQ
Sbjct: 586 GKLDLLSKMLKQLKADNHRVLLFSQMTKMLNVLEHFLEGE--GYQYDRIDGSIKGDLRQK 643
Query: 682 AIARFNQDKS-RFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
AI RFN S FVFLLSTR+ GLGINLATADTVII+DSD+NPH D+QA +RAHR+GQ
Sbjct: 644 AIDRFNDPVSEHFVFLLSTRAGGLGINLATADTVIIFDSDWNPHNDVQAFSRAHRMGQKK 703
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLDQLF--KGKSG-----SQKEVEDILKWGTEELF 793
++++YR V SVEERI+Q+AK K+ML L G G S+ E+EDIL++GTE+LF
Sbjct: 704 KVMIYRFVTHNSVEERIMQVAKHKMMLTHLVVRPGMGGMTTNFSKDELEDILRFGTEDLF 763
Query: 794 NDSCALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAIL 853
D S I +D+ A+
Sbjct: 764 KDG--------------------------------------------KSEAIHYDDKAVA 779
Query: 854 KLLDRSNLQDASTDIAEGDS-ENDMLGSMKALEW-NDEPTEEHVEGESPPHGADDMCTQN 911
LLDR+N I E +S N+ L S K + E EEH + + D +N
Sbjct: 780 DLLDRTN-----RGIEEKESWANEYLSSFKVASYATKEDHEEHDDYNNDAENTDPFYWEN 834
>CHD1_YEAST (P32657) Chromo domain protein 1
Length = 1468
Score = 464 bits (1195), Expect = e-130
Identities = 339/966 (35%), Positives = 505/966 (52%), Gaps = 96/966 (9%)
Query: 10 EDNSACDNDLDGEIAENLVHDPQNVKSSDEGELHN-TDRVEKIHVYRRSITKESKNGNLL 68
ED D+ I +N KS + + + +++ + + R +++K N
Sbjct: 79 EDFEEDDDYYGSPIKQNRSKPKSRTKSKSKSKPKSQSEKQSTVKIPTRFSNRQNKTVNY- 137
Query: 69 NSLSKATDDLGSCARDGTDQDDYAVSDEQLEKENDKLETEENLNVVLRGDGNSKLPNNCE 128
++ + DDL +DDY S+E L +EN E + N + N
Sbjct: 138 -NIDYSDDDLLE------SEDDYG-SEEALSEENVH---EASANPQPEDFHGIDIVINHR 186
Query: 129 MHDSLETKQKEVVLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNSWIS-ESHLK 187
+ SLE + VLEK V D + +YEFL+KW +SH+HN+W + ES +
Sbjct: 187 LKTSLEEGK---VLEK--------TVPDLNNCKENYEFLIKWTDESHLHNTWETYESIGQ 235
Query: 188 VIAKRKLENYKAK------------YGTAT-INICE----------EQWKNPERLLAIRT 224
V ++L+NY + Y TA I I + E++ PER++ +
Sbjct: 236 VRGLKRLDNYCKQFIIEDQQVRLDPYVTAEDIEIMDMERERRLDEFEEFHVPERIIDSQR 295
Query: 225 S--KQGTSEA--FVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFETLTLEREASK-- 278
+ + GTS+ VKW Y+E TWE+ + +++ + + F RE SK
Sbjct: 296 ASLEDGTSQLQYLVKWRRLNYDEATWENATD-IVKLAPEQVKHFQ-------NRENSKIL 347
Query: 279 -ENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLG 337
+ S+ +S R L QP ++GG L QL +NW+ W K N ILADEMGLG
Sbjct: 348 PQYSSNYTSQRPR-FEKLSVQPPFIKGGELRDFQLTGINWMAFLWSKGDNGILADEMGLG 406
Query: 338 KTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAII 397
KT+ AFIS L F + + P +++VPL TM WL F WAPD+N + Y G K+R I
Sbjct: 407 KTVQTVAFISWLIFARRQNGPHIIVVPLSTMPAWLDTFEKWAPDLNCICYMGNQKSRDTI 466
Query: 398 RQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSES 457
R+YE++ ++P KKT KFNVLLT+YE +L D + + W+ + VDE HRLKN+ES
Sbjct: 467 REYEFY-TNPRAKGKKT--MKFNVLLTTYEYILKDRAELGSIKWQFMAVDEAHRLKNAES 523
Query: 458 KLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKV 517
L+ LNS +R+L+TGTPLQNN+ E+ L+NFL P F + D E +
Sbjct: 524 SLYESLNSFKVANRMLITGTPLQNNIKELAALVNFLMPGRFTIDQEIDFENQDEEQEEYI 583
Query: 518 DELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKG 577
+L + + P +LRRLKKD +++P KTER++ VELS +Q EYY+ +LTKNY L KG
Sbjct: 584 HDLHRRIQPFILRRLKKDVEKSLPSKTERILRVELSDVQTEYYKNILTKNYSALTAGAKG 643
Query: 578 IAQQSMLNIVMQLRKVCNHPYLIPGTEP------DSGSVEFLHEMR--IKASAKLTLLHS 629
S+LNI+ +L+K NHPYL E G + + +R I +S K+ LL
Sbjct: 644 -GHFSLLNIMNELKKASNHPYLFDNAEERVLQKFGDGKMTRENVLRGLIMSSGKMVLLDQ 702
Query: 630 MLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQ- 688
+L L K+GHRVLIFSQM ++LDIL DYL+I+ ++R+DG+V R+ +I FN
Sbjct: 703 LLTRLKKDGHRVLIFSQMVRMLDILGDYLSIK--GINFQRLDGTVPSAQRRISIDHFNSP 760
Query: 689 DKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLV 748
D + FVFLLSTR+ GLGINL TADTV+I+DSD+NP AD+QAM RAHRIGQ N ++VYRLV
Sbjct: 761 DSNDFVFLLSTRAGGLGINLMTADTVVIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLV 820
Query: 749 VRASVEERILQLAKKKLMLD------------QLFKGKSGSQKEVEDILKWGTEELFNDS 796
+ +VEE +L+ A+KK++L+ + K + E+ ILK+G +F +
Sbjct: 821 SKDTVEEEVLERARKKMILEYAIISLGVTDGNKYTKKNEPNAGELSAILKFGAGNMFTAT 880
Query: 797 CALNG-KDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKL 855
+D + ++ N E GG + + + TD + I WD+ I+
Sbjct: 881 DNQKKLEDLNLDDVLNHAEDHVTTPDLGESHLGGEEFLKQFEVTDYKADIDWDD--IIPE 938
Query: 856 LDRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEEHVEGE--SPPHGADDMCTQNSE 913
+ LQD + + + L M + + + V G+ + +DD T S
Sbjct: 939 EELKKLQDEEQKRKDEEYVKEQLEMMNRRDNALKKIKNSVNGDGTAANSDSDDDSTSRSS 998
Query: 914 KKEDNA 919
++ A
Sbjct: 999 RRRARA 1004
>CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2
(CHD-2)
Length = 1739
Score = 464 bits (1193), Expect = e-130
Identities = 304/808 (37%), Positives = 452/808 (55%), Gaps = 114/808 (14%)
Query: 100 KENDKLETEENLNVVLRGDGNSKLPNNCEMHDSLETKQKEVVLEKGMGSSGDNKVQ---- 155
KE+D ET+ + + + G+G E D+ ET +K VL+ +G G
Sbjct: 233 KEDDDFETDSDDLIEMTGEGVD------EQQDNSETIEK--VLDSRLGKKGATGASTTVY 284
Query: 156 ------------DSIGEEVSYEFLVKWVGKSHIHNSWISESHL---KVIAKRKLENYKAK 200
D+ +E ++L+KW G S+IH++W SE L KV +KLEN+K K
Sbjct: 285 AIEANGDPSGDFDTEKDEGEIQYLIKWKGWSYIHSTWESEESLQQQKVKGLKKLENFKKK 344
Query: 201 -----------------YGTATINICEE---QWKNPERLLAIRTSKQ------------- 227
Y + E Q++ ER++A++TSK
Sbjct: 345 EDEIKQWLGKVSPEDVEYFNCQQELASELNKQYQIVERVIAVKTSKSTLGQTDFPAHSRK 404
Query: 228 ----GTSEAFVKWTGKPYNECTWESLDEPVL-QNSSHLITRFNMFETLTLEREASKENST 282
E KW G PY+EC+WE DE ++ + + I F+ R SK T
Sbjct: 405 PAPSNEPEYLCKWMGLPYSECSWE--DEALIGKKFQNCIDSFH-------SRNNSKTIPT 455
Query: 283 K--KSSDRQNDIVNLLEQPKELRGGSLF--PHQLEALNWLRKCWYKSRNVILADEMGLGK 338
+ K+ ++ V L +QP L G +L +QLE LNWL W K+ +VILADEMGLGK
Sbjct: 456 RECKALKQRPRFVALKKQPAYLGGENLELRDYQLEGLNWLAHSWCKNNSVILADEMGLGK 515
Query: 339 TISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIR 398
TI +F+S L+ + ++ P L++VPL T+ +W EF +WAP++NVV Y G +R IR
Sbjct: 516 TIQTISFLSYLFHQHQLYGPFLIVVPLSTLTSWQREFEIWAPEINVVVYIGDLMSRNTIR 575
Query: 399 QYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESK 458
+YEW ++ +T+ KFN L+T+YE++L D + + W L VDE HRLKN +S
Sbjct: 576 EYEW-------IHSQTKRLKFNALITTYEILLKDKTVLGSINWAFLGVDEAHRLKNDDSL 628
Query: 459 LFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVD 518
L+ L HR+L+TGTPLQN+L E+++LL+F+ P F FEE +
Sbjct: 629 LYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFEFWEDFEED-HGKGRENGYQ 687
Query: 519 ELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGI 578
L K++ P +LRR+KKD +++P K E+++ VE+S++Q +YY+ +LT+NY+ L +G
Sbjct: 688 SLHKVLEPFLLRRVKKDVEKSLPAKVEQILRVEMSALQKQYYKWILTRNYKALAKGTRG- 746
Query: 579 AQQSMLNIVMQLRKVCNHPYLI-PGTEPDSGSVEFLHEMRIKASAKLTLLHSMLKILYKE 637
+ LNIVM+L+K CNH YLI P E + + + + I++S KL LL +L L +
Sbjct: 747 STSGFLNIVMELKKCCNHCYLIKPPEENERENGQEILLSLIRSSGKLILLDKLLTRLRER 806
Query: 638 GHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKSR-FVFL 696
G+RVLIFSQM ++LDIL +YL I+ P ++R+DGS+ R+ A+ FN D S F FL
Sbjct: 807 GNRVLIFSQMVRMLDILAEYLTIKHYP--FQRLDGSIKGEIRKQALDHFNADGSEDFCFL 864
Query: 697 LSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEER 756
LSTR+ GLGINLA+ADTV+I+DSD+NP D+QA RAHRIGQ ++ +YRLV + +VEE
Sbjct: 865 LSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEE 924
Query: 757 ILQLAKKKLMLDQLF---------------KGKSGS----QKEVEDILKWGTEELFNDSC 797
I++ AKKK++LD L G+S S ++E+ ILK+G E+LF +
Sbjct: 925 IIERAKKKMVLDHLVIQRMDTTGRTILENNSGRSNSNPFNKEELTAILKFGAEDLFKE-- 982
Query: 798 ALNGKDTSENNNSNKDEAVAEVEHKHRK 825
L G++ SE + DE + E + +
Sbjct: 983 -LEGEE-SEPQEMDIDEILRLAETRENE 1008
>CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7
(CHD-7) (Fragment)
Length = 1967
Score = 462 bits (1188), Expect = e-129
Identities = 288/705 (40%), Positives = 407/705 (56%), Gaps = 60/705 (8%)
Query: 124 PNNCEMHDSLETKQKE-VVLEKGMGSSGDNKVQDSIGEEVSYE-FLVKWVGKSHIHNSWI 181
P+N + E+ E V+EK M S K ++S GEEV E F VK+ S++H W
Sbjct: 21 PSNTSQSEQQESVDAEGPVVEKIMSSRSVKKQKES-GEEVEIEEFYVKYKNFSYLHCQWA 79
Query: 182 SESHLKVIAK--RKLENYKAKYG-TATINICEEQWKNPE-----RLLAIRTSKQGTSEA- 232
S L+ + +K++ +KAK G ++ E++ NP+ R++ S E
Sbjct: 80 SIEDLEKDKRIQQKIKRFKAKQGQNKFLSEIEDELFNPDYVEVDRIMDFARSTDDRGEPV 139
Query: 233 ---FVKWTGKPYNECTWE---SLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSS 286
VKW PY + TWE +D+ ++ L++R ET +ER + ++ K S
Sbjct: 140 THYLVKWCSLPYEDSTWERRQDIDQAKIEEFEKLMSREP--ETERVERPPA-DDWKKSES 196
Query: 287 DRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFI 346
R+ N L + +QLE +NWL WY RN ILADEMGLGKTI + F+
Sbjct: 197 SREYKNNNKLRE-----------YQLEGVNWLLFNWYNMRNCILADEMGLGKTIQSITFL 245
Query: 347 SSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASD 406
+Y + + P LV+ PL T+ NW EF W ++NVV YHG +R I+ YE + D
Sbjct: 246 YEIYLK-GIHGPFLVIAPLSTIPNWEREFRTWT-ELNVVVYHGSQASRRTIQLYEMYFKD 303
Query: 407 PSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSI 466
P G K +YKF+ ++T++EM+L D R +PW +++DE HRLKN KL L +
Sbjct: 304 PQGRVIKG-SYKFHAIITTFEMILTDCPELRNIPWRCVVIDEAHRLKNRNCKLLEGLKMM 362
Query: 467 SFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSP 526
+H+VLLTGTPLQN + E+++LL+FL+P+ FPS + F + F DL + E+V +L+ ++ P
Sbjct: 363 DLEHKVLLTGTPLQNTVEELFSLLHFLEPSRFPSETTFMQEFGDLKTEEQVQKLQAILKP 422
Query: 527 HMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNI 586
MLRRLK+D +N+ PK E ++ VEL++IQ +YYRA+L KN+ L G ++LN
Sbjct: 423 MMLRRLKEDVEKNLAPKEETIIEVELTNIQKKYYRAILEKNFTFLSKGGGQANVPNLLNT 482
Query: 587 VMQLRKVCNHPYLIPGTEP----------DSGSVEFLHEMRIKASAKLTLLHSMLKILYK 636
+M+LRK CNHPYLI G E ++ S +F + I+A+ KL L+ +L L
Sbjct: 483 MMELRKCCNHPYLINGAEEKILEEFKETHNAESPDFQLQAMIQAAGKLVLIDKLLPKLKA 542
Query: 637 EGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQ-DKSRFVF 695
GHRVLIFSQM + LDILEDYL P YER+DG V RQ AI RF++ D RFVF
Sbjct: 543 GGHRVLIFSQMVRCLDILEDYLIQRRYP--YERIDGRVRGNLRQAAIDRFSKPDSDRFVF 600
Query: 696 LLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEE 755
LL TR+ GLGINL ADT II+DSD+NP D+QA R HRIGQS + +YRL+ R S E
Sbjct: 601 LLCTRAGGLGINLTAADTCIIFDSDWNPQNDLQAQARCHRIGQSKSVKIYRLITRNSYER 660
Query: 756 RILQLAKKKLMLDQ-LFKGKSG-----------SQKEVEDILKWG 788
+ A KL LD+ + + SG S+KE+ED+L+ G
Sbjct: 661 EMFDKASLKLGLDKAVLQSMSGRENATNGVQQLSKKEIEDLLRKG 705
>HRP3_SCHPO (O14139) Chromodomain helicase hrp3
Length = 1388
Score = 453 bits (1166), Expect = e-127
Identities = 308/900 (34%), Positives = 473/900 (52%), Gaps = 100/900 (11%)
Query: 37 SDEGELHNTDRVEKIHVYRRSITKESKNGNLLN---SLSKATDDLGSCARDGTDQDDYAV 93
S+ G +TD E V+ K ++NG+ + ++ D+ S + ++ DD +
Sbjct: 69 SNPGRYVDTDDQED--VFPSKHRKGTRNGSSFSRHRTIRDLDDEAESVTSEESESDDSSY 126
Query: 94 SDEQLEKENDKLETEENLNVVLRGDGNSKLPNNCE--MHDSLETKQKEVVLEKGMGSSG- 150
++ K + + NSK N E +S E +++E + E S
Sbjct: 127 GGTPKKRSRQKKSNTYVQDEIRFSSRNSKGVNYNEDAYFESFEEEEEEEMYEYATEVSEE 186
Query: 151 --DNKVQDSI-------GEEVS-----YEFLVKWVGKSHIHNSWISESHLKVI-AKRKLE 195
D + D + G + S YEFL+KWV SH+H +W +++ +I +K++
Sbjct: 187 PEDTRAIDVVLDHRLIEGHDGSTPSEDYEFLIKWVNFSHLHCTWEPYNNISMIRGSKKVD 246
Query: 196 NYKAKYGTATINICE-----------------------EQWKNPERLLAIRTSKQGTSEA 232
N+ + I E E++K +R++A + G+ E
Sbjct: 247 NHIKQVILLDREIREDPTTTREDIEAMDIEKERKRENYEEYKQVDRIVAKHLNSDGSVEY 306
Query: 233 FVKWTGKPYNECTWE--SLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSS--DR 288
VKW Y+ CTWE S+ EP+ T F+ ERE S + ++ ++ +
Sbjct: 307 LVKWKQLLYDFCTWEASSIIEPI------AATEIQAFQ----EREESALSPSRGTNYGNS 356
Query: 289 QNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISS 348
+ L +QP + GG L QL +NW+ W+K+ N ILADEMGLGKT+ AF+S
Sbjct: 357 RPKYRKLEQQPSYITGGELRDFQLTGVNWMAYLWHKNENGILADEMGLGKTVQTVAFLSY 416
Query: 349 LYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPS 408
L + P LV+VPL T+ W ALWA D+N + Y G +R +IR YE++
Sbjct: 417 LAHSLRQHGPFLVVVPLSTVPAWQETLALWASDMNCISYLGNTTSRQVIRDYEFYVDG-- 474
Query: 409 GLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISF 468
T+ KFN+LLT+YE VL D S + W+ + +DE HRLKNSES L+ L+
Sbjct: 475 -----TQKIKFNLLLTTYEYVLKDRSVLSNIKWQYMAIDEAHRLKNSESSLYEALSQFKN 529
Query: 469 QHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHM 528
+R+L+TGTPLQNN+ E+ L++FL P F D + L++ + P++
Sbjct: 530 SNRLLITGTPLQNNIRELAALVDFLMPGKFEIREEINLEAPDEEQEAYIRSLQEHLQPYI 589
Query: 529 LRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVM 588
LRRLKKD +++P K+ER++ VELS +Q +Y+ +LT+NY++L +Q S+LNIV+
Sbjct: 590 LRRLKKDVEKSLPSKSERILRVELSDLQMYWYKNILTRNYRVLTQSISSGSQISLLNIVV 649
Query: 589 QLRKVCNHPYLIPGTEP-------DSGSVEFLHEMRIKASAKLTLLHSMLKILYKEGHRV 641
+L+K NHPYL G E G + + + I S K+ LL +L L ++GHRV
Sbjct: 650 ELKKASNHPYLFDGVEESWMQKINSQGRRDEVLKGLIMNSGKMVLLDKLLSRLRRDGHRV 709
Query: 642 LIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQDKS-RFVFLLSTR 700
LIFSQM ++LDIL DYL++ P ++R+DG+V R+T+I FN S FVFLLSTR
Sbjct: 710 LIFSQMVRMLDILGDYLSLRGYP--HQRLDGTVPAAVRRTSIDHFNAPNSPDFVFLLSTR 767
Query: 701 SCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQL 760
+ GLGINL TADTVII+DSD+NP AD+QAM RAHRIGQ N ++VYRL+ + ++EE +L+
Sbjct: 768 AGGLGINLMTADTVIIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLLSKDTIEEDVLER 827
Query: 761 AKKKLMLDQLF------------KGKSGSQKEVEDILKWGTEELFNDSCALNGKDTSENN 808
A++K++L+ K S +E+ ILK+G +F A N + E+
Sbjct: 828 ARRKMILEYAIISLGVTDKQKNSKNDKFSAEELSAILKFGASNMFK---AENNQKKLEDM 884
Query: 809 NSNKDEAVAEVEHKHRKRTGGLGDVYED-----KCTDNSSKIMWDENAILKLLDRSNLQD 863
N ++ AE +H GG E+ + TD + + WD+ I+ L +R ++
Sbjct: 885 NLDEILEHAE-DHDTSNDVGGASMGGEEFLKQFEVTDYKADVSWDD--IIPLTEREKFEE 941
>HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1
Length = 1373
Score = 453 bits (1165), Expect = e-126
Identities = 294/844 (34%), Positives = 446/844 (52%), Gaps = 94/844 (11%)
Query: 119 GNSKLPNNCEMHDSLETKQKEV----------VLEKGMGSSGDNKVQDSIGEEVSYEFLV 168
G S N E +D E +++EV +++ + QD + SY++L+
Sbjct: 176 GKSVNYNEQEFYDDFEDEEEEVEEQVEEEYEPIIDFVLNHRKRADAQDD-DPKSSYQYLI 234
Query: 169 KWVGKSHIHNSWISESHLKVI-AKRKLENYKAKY----------GTATI----------- 206
KW SH+HN+W S L + +K++NY + T T
Sbjct: 235 KWQEVSHLHNTWEDYSTLSSVRGYKKVDNYIKQNIIYDREIREDPTTTFEDIEALDIERE 294
Query: 207 --NICEEQWKNPERLLAIRTSKQGTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRF 264
N+ E++K ER++A T+++G +E FVKW PY+ CTWE D + + + + +F
Sbjct: 295 RKNMLFEEYKIVERIVASETNEEGKTEYFVKWRQLPYDNCTWEDADV-IYSMAPNEVYQF 353
Query: 265 NMFETLTLEREASKENSTKKSSDRQNDIVNLLE-QPKELRGGSLFPHQLEALNWLRKCWY 323
L+RE S K LE QP ++GG + QL +NW+ W+
Sbjct: 354 -------LQRENSPYLPYKGVFYNTRPPYRKLEKQPSYIKGGEIRDFQLTGINWMAYLWH 406
Query: 324 KSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVN 383
++ N ILADEMGLGKT+ F+S L K P L++VPL T+ W A W PD+N
Sbjct: 407 RNENGILADEMGLGKTVQTVCFLSYLVHSLKQHGPFLIVVPLSTVPAWQETLANWTPDLN 466
Query: 384 VVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEV 443
+ Y G ++RA IR+YE++ L+ + KFN+LLT+YE +L D + W+
Sbjct: 467 SICYTGNTESRANIREYEFY------LSTNSRKLKFNILLTTYEYILKDKQELNNIRWQY 520
Query: 444 LIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSA 503
L +DE HRLKNSES L+ L+ +R+L+TGTPLQNNL E+ +L+NFL P F
Sbjct: 521 LAIDEAHRLKNSESSLYETLSQFRTANRLLITGTPLQNNLKELASLVNFLMPGKFYIRDE 580
Query: 504 FEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAM 563
+ + +L++ + P +LRRLKKD +++P K+ER++ VELS +Q E+Y+ +
Sbjct: 581 LNFDQPNAEQERDIRDLQERLQPFILRRLKKDVEKSLPSKSERILRVELSDMQTEWYKNI 640
Query: 564 LTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGT-EPDSGSVEFLHEMRIKA-- 620
LTKNY+ L G Q S+LNIV++L+KV NHPYL PG E + E ++
Sbjct: 641 LTKNYRALTGHTDGRGQLSLLNIVVELKKVSNHPYLFPGAAEKWMMGRKMTREDTLRGII 700
Query: 621 --SAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTD 678
S K+ LL +L+ L +GHRVLIFSQM ++L+IL +Y+++ Y+R+DG++ +
Sbjct: 701 MNSGKMVLLDKLLQRLKHDGHRVLIFSQMVRMLNILGEYMSLR--GYNYQRLDGTIPASV 758
Query: 679 RQTAIARFNQ-DKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIG 737
R+ +I FN D FVFLLSTR+ GLGINL TADTVII+DSD+NP AD+QAM RAHRIG
Sbjct: 759 RRVSIDHFNAPDSPDFVFLLSTRAGGLGINLNTADTVIIFDSDWNPQADLQAMARAHRIG 818
Query: 738 QSNRLLVYRLVVRASVEERILQLAKKKLMLDQLF------------KGKSGSQKEVEDIL 785
Q N + VYR + + +VEE IL+ A++K++L+ K +E+ IL
Sbjct: 819 QKNHVNVYRFLSKDTVEEDILERARRKMILEYAIISLGVTEKSKNSKNDKYDAQELSAIL 878
Query: 786 KWGTEELFNDSCALNGKDTSENNNSNKDEAVAEVEHKHRKR------TGGLGDVYEDKCT 839
K+G +F + ++ + N N D+ ++ E + GG + + + T
Sbjct: 879 KFGASNMFKAT-----ENQKKLENMNLDDILSHAEDRDSSNDVGGASMGGEEFLKQFEVT 933
Query: 840 D-NSSKIMWDENAILKLLDRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEEHVEGE 898
D + + WD+ + ++R + E ML + +A E E EE
Sbjct: 934 DYKAEDLNWDDIIPEEEMERI------------EEEERMLAAQRAKEEERERREEEEREN 981
Query: 899 SPPH 902
H
Sbjct: 982 DEDH 985
>CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1711
Score = 436 bits (1120), Expect = e-121
Identities = 303/860 (35%), Positives = 459/860 (53%), Gaps = 102/860 (11%)
Query: 10 EDNSACDNDLDGEIAENLVHDPQNVKSSDEGELHNTDRVEKIHVYRRSITKESKNGNLLN 69
ED + ++ + + + KSS +G + + K+ + +SKNG +
Sbjct: 146 EDWQMSGSGSPSQLGSDSESEEERDKSSCDGTESDYEPKNKVRSRKPQNRSKSKNGKKIL 205
Query: 70 SLSKATDDLGSCARDGTDQDDYAVSDEQLE-----KENDKLETEENLNVVLRGDGNSKLP 124
K D D +D S Q KE+++++T+ + + + G+ +
Sbjct: 206 GQKKRQIDSSEDEDDEDYDNDKRSSRRQATVNVSYKEDEEMKTDSDDLLEVCGEDVPQPE 265
Query: 125 NNCEMHDSLETKQKEVVLEKG----------MGSSGD-NKVQDSIGEEVSYEFLVKWVGK 173
+ E +++E V KG + + GD N + E ++L+KW G
Sbjct: 266 D--EEFETIERVMDCRVGRKGATGATTTIYAVEADGDPNAGFERNKEPGDIQYLIKWKGW 323
Query: 174 SHIHNSWISESHLK---VIAKRKLENYKAK-----------------YGTATINICEE-- 211
SHIHN+W +E LK V +KL+NYK K Y + ++
Sbjct: 324 SHIHNTWETEETLKQQNVRGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELTDDLH 383
Query: 212 -QWKNPERLLAIRTSKQ--GTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFE 268
Q++ ER++A K G + + KW G PY+EC+WE LI++ F+
Sbjct: 384 KQYQIVERIIAHSNQKSAAGLPDYYCKWQGLPYSECSWED---------GALISK--KFQ 432
Query: 269 TLTLEREASKENSTKKSSD-----RQNDIVNLLEQPKELRGGS---LFPHQLEALNWLRK 320
T E + ++ T D ++ V L +QP + G L +QL LNWL
Sbjct: 433 TCIDEYFSRNQSKTTPFKDCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAH 492
Query: 321 CWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAP 380
W K + ILADEMGLGKTI +F++ L+ E ++ P L++VPL T+ +W E WA
Sbjct: 493 SWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTWAS 552
Query: 381 DVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVP 440
+N V Y G +R +IR +EW ++ +T+ KFN+LLT+YE++L D + G+
Sbjct: 553 QMNAVVYLGDINSRNMIRTHEW-------MHPQTKRLKFNILLTTYEILLKDKAFLGGLN 605
Query: 441 WEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPS 500
W + VDE HRLKN +S L+ L HR+L+TGTPLQN+L E+++LL+F+ P F S
Sbjct: 606 WAFIGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSS 665
Query: 501 LSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYY 560
FEE L K + P +LRR+KKD +++P K E+++ +E+S++Q +YY
Sbjct: 666 WEDFEEEHGKGREYGYAS-LHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYY 724
Query: 561 RAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMR--- 617
+ +LT+NY+ L KG + LNI+M+L+K CNH YLI PD+ EF ++
Sbjct: 725 KWILTRNYKALSKGSKG-STSGFLNIMMELKKCCNHCYLIK--PPDNN--EFYNKQEALQ 779
Query: 618 --IKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVS 675
I++S KL LL +L L + G+RVLIFSQM ++LDIL +YL P ++R+DGS+
Sbjct: 780 HLIRSSGKLILLDKLLIRLRERGNRVLIFSQMVRMLDILAEYLKYRQFP--FQRLDGSIK 837
Query: 676 VTDRQTAIARFNQDKSR-FVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAH 734
R+ A+ FN + S F FLLSTR+ GLGINLA+ADTV+I+DSD+NP D+QA RAH
Sbjct: 838 GELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAH 897
Query: 735 RIGQSNRLLVYRLVVRASVEERILQLAKKKLMLDQLF------KGK----SGS------- 777
RIGQ ++ +YRLV + SVEE IL+ AKKK++LD L GK +GS
Sbjct: 898 RIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVIQRMDTTGKTVLHTGSAPSSSTP 957
Query: 778 --QKEVEDILKWGTEELFND 795
++E+ ILK+G EELF +
Sbjct: 958 FNKEELSAILKFGAEELFKE 977
>CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8
(CHD-8) (Helicase with SNF2 domain 1) (Fragment)
Length = 2004
Score = 432 bits (1112), Expect = e-120
Identities = 279/747 (37%), Positives = 409/747 (54%), Gaps = 66/747 (8%)
Query: 86 TDQDDYAVSDEQLEKEND---KLETEENLNVVLRGDGNSKLPNNCEMHDSLETKQKE--V 140
T+ D ++D++ E+E D ++ E L ++ LP+ M +E +E
Sbjct: 9 TEDLDIKITDDEEEEEVDVTGPIKPEPILPEPVQEPDGETLPS---MQFFVENPSEEDAA 65
Query: 141 VLEKGMGSSGDNKVQDSIGEEVSYEFLVKWVGKSHIHNSWISESHLKVIAK--RKLENYK 198
+++K + K S + EF VK+ S++H W + S L+ + +KL+ +K
Sbjct: 66 IVDKVLSMRIVKKELPSGQYTEAEEFFVKYKNYSYLHCEWATISQLEKDKRIHQKLKRFK 125
Query: 199 AKYGTATINICEEQWK-NPERLLAIRTSKQGTS----------EAFVKWTGKPYNECTWE 247
K E++ NP+ + R + S VKW PY + TWE
Sbjct: 126 TKMAQMRHFFHEDEEPFNPDYVEVDRILDESHSIDKDNGEPVIYYLVKWCSLPYEDSTWE 185
Query: 248 ---SLDEPVLQNSSHLITRFNMFETLTLEREASKENSTKKSSDRQNDIVNLLEQPKELRG 304
+DE ++ + +R E + N + S+ ++ ++ + + +LR
Sbjct: 186 LKEDVDEGKIREFKRIQSR---------HPELKRVNRPQASAWKKLELSHEYKNRNQLR- 235
Query: 305 GSLFPHQLEALNWLRKCWYKSRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVP 364
+QLE +NWL WY +N ILADEMGLGKTI + AF+ +Y + P LV+ P
Sbjct: 236 ----EYQLEGVNWLLFNWYNRQNCILADEMGLGKTIQSIAFLQEVY-NVGIHGPFLVIAP 290
Query: 365 LVTMGNWLAEFALWAPDVNVVQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLT 424
L T+ NW EF W ++N + YHG +R +I+QYE + D G AYKF+ L+T
Sbjct: 291 LSTITNWEREFNTWT-EMNTIVYHGSLASRQMIQQYEMYCKDSRG-RLIPGAYKFDALIT 348
Query: 425 SYEMVLADYSHFRGVPWEVLIVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLG 484
++EM+L+D R + W +I+DE HRLKN KL L + +H+VLLTGTPLQN +
Sbjct: 349 TFEMILSDCPELREIEWRCVIIDEAHRLKNRNCKLLDSLKHMDLEHKVLLTGTPLQNTVE 408
Query: 485 EMYNLLNFLQPASFPSLSAFEERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKT 544
E+++LL+FL+P+ FPS S F + F DL + E+V +L+ ++ P MLRRLK+D +N+ PK
Sbjct: 409 ELFSLLHFLEPSQFPSESEFLKDFGDLKTEEQVQKLQAILKPMMLRRLKEDVEKNLAPKQ 468
Query: 545 ERMVPVELSSIQAEYYRAMLTKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTE 604
E ++ VEL++IQ +YYRA+L KN+ L ++LN +M+LRK CNHPYLI G E
Sbjct: 469 ETIIEVELTNIQKKYYRAILEKNFSFLSKGAGHTNMPNLLNTMMELRKCCNHPYLINGAE 528
Query: 605 PDSGSV----------EFLHEMRIKASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDIL 654
+ +F + ++++ KL L+ +L L GH+VLIFSQM + LDIL
Sbjct: 529 EKILTEFREACHIIPHDFHLQAMVRSAGKLVLIDKLLPKLKAGGHKVLIFSQMVRCLDIL 588
Query: 655 EDYLNIEFGPKTYERVDGSVSVTDRQTAIARFNQ-DKSRFVFLLSTRSCGLGINLATADT 713
EDYL YER+DG V RQ AI RF++ D RFVFLL TR+ GLGINL ADT
Sbjct: 589 EDYLIQR--RYLYERIDGRVRGNLRQAAIDRFSKPDSDRFVFLLCTRAGGLGINLTAADT 646
Query: 714 VIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYRLVVRASVEERILQLAKKKLMLD----Q 769
II+DSD+NP D+QA R HRIGQS + VYRL+ R S E + A KL LD Q
Sbjct: 647 CIIFDSDWNPQNDLQAQARCHRIGQSKAVKVYRLITRNSYEREMFDKASLKLGLDKAVLQ 706
Query: 770 LFKGKSG--------SQKEVEDILKWG 788
G+ G S+KE+ED+L+ G
Sbjct: 707 SMSGRDGNITGIQQFSKKEIEDLLRKG 733
>CHD6_HUMAN (Q8TD26) Chromodomain-helicase-DNA-binding protein 6
(CHD-6)
Length = 2713
Score = 431 bits (1109), Expect = e-120
Identities = 270/662 (40%), Positives = 376/662 (56%), Gaps = 59/662 (8%)
Query: 166 FLVKWVGKSHIHNSWISESHLKVIAK--RKLENYKAKYGTATINICE--EQWKNPE---- 217
F VK+ S++H W + L+ + +K++ ++ K E E NP+
Sbjct: 316 FYVKYRNFSYLHCKWATMEELEKDPRIAQKIKRFRNKQAQMKHIFTEPDEDLFNPDYVEV 375
Query: 218 -RLLAIRTSKQG-----TSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFETLT 271
R+L + +K + VKW PY E TWE L+E V + FE+L
Sbjct: 376 DRILEVAHTKDAETGEEVTHYLVKWCSLPYEESTWE-LEEDVDP------AKVKEFESLQ 428
Query: 272 LEREASKENSTKKSSDRQNDIVNLLEQPKELRGGS-LFPHQLEALNWLRKCWYKSRNVIL 330
+ E K +D LE+ +E + + L +QLE +NWL WY +N IL
Sbjct: 429 VLPEI------KHVERPASDSWQKLEKSREYKNSNQLREYQLEGMNWLLFNWYNRKNCIL 482
Query: 331 ADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGC 390
ADEMGLGKTI + F+S ++ + P L++ PL T+ NW EF W ++N + YHG
Sbjct: 483 ADEMGLGKTIQSITFLSEIFLR-GIHGPFLIIAPLSTITNWEREFRTWT-EMNAIVYHGS 540
Query: 391 AKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGH 450
+R +I+QYE D G N + +KF+V++T++EM+LAD + + W +I+DE H
Sbjct: 541 QISRQMIQQYEMVYRDAQG-NPLSGVFKFHVVITTFEMILADCPELKKIHWSCVIIDEAH 599
Query: 451 RLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFND 510
RLKN KL L ++ +H+VLLTGTPLQN++ E+++LLNFL+P+ FPS +AF E F D
Sbjct: 600 RLKNRNCKLLEGLKLMALEHKVLLTGTPLQNSVEELFSLLNFLEPSQFPSETAFLEEFGD 659
Query: 511 LTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNYQI 570
L + E+V +L+ ++ P MLRRLK D +N+ PK E ++ VEL++IQ +YYRA+L KN+
Sbjct: 660 LKTEEQVKKLQSILKPMMLRRLKDDVEKNLAPKQETIIEVELTNIQKKYYRAILEKNFSF 719
Query: 571 LRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTE------------PDSGSVEFLHEMRI 618
L +++N +M+LRK CNHPYLI G E PD + +F + I
Sbjct: 720 LTKGANQHNMPNLINTMMELRKCCNHPYLINGAEEKILEDFRKTHSPD--APDFQLQAMI 777
Query: 619 KASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTD 678
+A+ KL L+ +L L GH+VLIFSQM + LDILEDYL TYER+DG V
Sbjct: 778 QAAGKLVLIDKLLPKLIAGGHKVLIFSQMVRCLDILEDYLIQR--RYTYERIDGRVRGNL 835
Query: 679 RQTAIARF-NQDKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIG 737
RQ AI RF D RFVFLL TR+ GLGINL ADT II+DSD+NP D+QA R HRIG
Sbjct: 836 RQAAIDRFCKPDSDRFVFLLCTRAGGLGINLTAADTCIIFDSDWNPQNDLQAQARCHRIG 895
Query: 738 QSNRLLVYRLVVRASVEERILQLAKKKLMLD----QLFKGKSG-------SQKEVEDILK 786
QS + VYRL+ R S E + A KL LD Q K G S+ EVED+L+
Sbjct: 896 QSKAVKVYRLITRNSYEREMFDKASLKLGLDKAVLQDINRKGGTNGVQQLSKMEVEDLLR 955
Query: 787 WG 788
G
Sbjct: 956 KG 957
>CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1709
Score = 431 bits (1107), Expect = e-120
Identities = 301/856 (35%), Positives = 451/856 (52%), Gaps = 100/856 (11%)
Query: 2 SENQTRLAEDNSACDNDLDGEIAENLVHD--PQNVKSSDEGELHNTDRVEKIHVYRRSIT 59
S++++ + S+CD +N V PQN S G+ + +I
Sbjct: 162 SDSESEEEREKSSCDETESDYEPKNKVKSRKPQNRSKSKNGKKILGQKKRQIDSSEEDDD 221
Query: 60 KESKNGNLLNSLSKATDDLGSCARDG--TDQDDY--AVSDEQLEKENDKLETEENLNVVL 115
+E + + +S +AT ++ + TD DD ++ + E ++ ET E
Sbjct: 222 EEDYDNDKRSSRRQATVNVSYKEDEEMKTDSDDLLEVCGEDVPQPEEEEFETIERFM--- 278
Query: 116 RGDGNSKLPNNCEMHDSLETKQKEVVLEKGMGSSGD-NKVQDSIGEEVSYEFLVKWVGKS 174
+C + T + + + GD N + E ++L+KW G S
Sbjct: 279 ----------DCRIGRKGATGATTTIY--AVEADGDPNAGFEKNKEPGEIQYLIKWKGWS 326
Query: 175 HIHNSWISESHLK---VIAKRKLENYKAK-----------------YGTATINICEE--- 211
HIHN+W +E LK V +KL+NYK K Y + ++
Sbjct: 327 HIHNTWETEETLKQQNVRGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELTDDLHK 386
Query: 212 QWKNPERLLAIRTSKQ--GTSEAFVKWTGKPYNECTWESLDEPVLQNSSHLITRFNMFET 269
Q++ R++A K G + + KW G PY+EC+WE + + + +F
Sbjct: 387 QYQIVGRIIAHSNQKSAAGYPDYYCKWQGLPYSECSWE--------DGALISKKFQACID 438
Query: 270 LTLEREASKENSTK--KSSDRQNDIVNLLEQPKELRGGS---LFPHQLEALNWLRKCWYK 324
R SK K K ++ V L +QP + G L +QL LNWL W K
Sbjct: 439 EYFSRNQSKTTPFKDCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCK 498
Query: 325 SRNVILADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNV 384
+ ILADEMGLGKTI +F++ L+ E ++ P L++VPL T+ +W E WA +N
Sbjct: 499 GNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTWASQMNA 558
Query: 385 VQYHGCAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVL 444
V Y G +R +IR +EW + +T+ KFN+LLT+YE++L D + G+ W +
Sbjct: 559 VVYLGDINSRNMIRTHEW-------THHQTKRLKFNILLTTYEILLKDKAFLGGLNWAFI 611
Query: 445 IVDEGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAF 504
VDE HRLKN +S L+ L HR+L+TGTPLQN+L E+++LL+F+ P F S F
Sbjct: 612 GVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDF 671
Query: 505 EERFNDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAML 564
EE L K + P +LRR+KKD +++P K E+++ +E+S++Q +YY+ +L
Sbjct: 672 EEEHGKGREYGYAS-LHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWIL 730
Query: 565 TKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMR-----IK 619
T+NY+ L KG + LNI+M+L+K CNH YLI PD+ EF ++ I+
Sbjct: 731 TRNYKALSKGSKG-STSGFLNIMMELKKCCNHCYLIK--PPDNN--EFYNKQEALQHLIR 785
Query: 620 ASAKLTLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDR 679
+S KL LL +L L + G+RVLIFSQM ++LDIL +YL P ++R+DGS+ R
Sbjct: 786 SSGKLILLDKLLIRLRERGNRVLIFSQMVRMLDILAEYLKYRQFP--FQRLDGSIKGELR 843
Query: 680 QTAIARFNQDKSR-FVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQ 738
+ A+ FN + S F FLLSTR+ GLGINLA+ADTV+I+DSD+NP D+QA RAHRIGQ
Sbjct: 844 KQALDHFNAEGSEDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQ 903
Query: 739 SNRLLVYRLVVRASVEERILQLAKKKLMLDQLF------KGK----SGS---------QK 779
++ +YRLV + SVEE IL+ AKKK++LD L GK +GS ++
Sbjct: 904 KKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVIQRMDTTGKTVLHTGSAPSSSTPFNKE 963
Query: 780 EVEDILKWGTEELFND 795
E+ ILK+G EELF +
Sbjct: 964 ELSAILKFGAEELFKE 979
>ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain ISW1
(EC 3.6.1.-)
Length = 1129
Score = 375 bits (963), Expect = e-103
Identities = 231/595 (38%), Positives = 333/595 (55%), Gaps = 35/595 (5%)
Query: 273 EREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILAD 332
E E E ++ SD I + G L P+Q++ +NWL ILAD
Sbjct: 162 EHEEDAELLKEEDSDDDESIEFQFRESPAYVNGQLRPYQIQGVNWLVSLHKNKIAGILAD 221
Query: 333 EMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAK 392
EMGLGKT+ +F+ L + K+ P LV+ P T+ NWL E W PDVN G +
Sbjct: 222 EMGLGKTLQTISFLGYLRYIEKIPGPFLVIAPKSTLNNWLREINRWTPDVNAFILQGDKE 281
Query: 393 ARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEGHRL 452
RA + Q KK F+V++ SYE+++ + S + + WE +I+DE HR+
Sbjct: 282 ERAELIQ------------KKLLGCDFDVVIASYEIIIREKSPLKKINWEYIIIDEAHRI 329
Query: 453 KNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFNDLT 512
KN ES L +L + ++R+L+TGTPLQNNL E++ LLNFL P F F++ F+ +
Sbjct: 330 KNEESMLSQVLREFTSRNRLLITGTPLQNNLHELWALLNFLLPDIFSDAQDFDDWFSSES 389
Query: 513 SAEKVDELKK----LVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTKNY 568
+ E D++ K ++ P +LRR+K D ++ PK E + V +SS+Q ++Y+ +L K+
Sbjct: 390 TEEDQDKIVKQLHTVLQPFLLRRIKSDVETSLLPKKELNLYVGMSSMQKKWYKKILEKDL 449
Query: 569 QILRNI-GKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTLL 627
+ G ++ +LNI+MQLRK CNHPYL G EP G E + +AKL +L
Sbjct: 450 DAVNGSNGSKESKTRLLNIMMQLRKCCNHPYLFDGAEP--GPPYTTDEHLVYNAAKLQVL 507
Query: 628 HSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARFN 687
+LK L +EG RVLIFSQM++LLDILEDY F Y R+DGS + DR AI +N
Sbjct: 508 DKLLKKLKEEGSRVLIFSQMSRLLDILEDYCY--FRNYEYCRIDGSTAHEDRIQAIDDYN 565
Query: 688 Q-DKSRFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVYR 746
D +FVFLL+TR+ GLGINL +AD V++YDSD+NP AD+QAM+RAHRIGQ ++ V+R
Sbjct: 566 APDSKKFVFLLTTRAGGLGINLTSADVVVLYDSDWNPQADLQAMDRAHRIGQKKQVKVFR 625
Query: 747 LVVRASVEERILQLAKKKLMLDQLF-----------KGKSGSQKEVEDILKWGTEELFND 795
LV SVEE+IL+ A +KL LDQL + K+ S+ + +++ G ++F
Sbjct: 626 LVTDNSVEEKILERATQKLRLDQLVIQQNRTSLKKKENKADSKDALLSMIQHGAADVFKS 685
Query: 796 SCALNGKDTSENNNSNK--DEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWD 848
+ T E + K D + E+ K +T L YE D+ K D
Sbjct: 686 GTSTGSAGTPEPGSGEKGDDIDLDELLLKSENKTKSLNAKYETLGLDDLQKFNQD 740
>ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPase
chain (EC 3.6.1.-) (ISW2-like) (Sucrose nonfermenting
protein 2 homolog)
Length = 1057
Score = 370 bits (950), Expect = e-102
Identities = 233/637 (36%), Positives = 348/637 (54%), Gaps = 57/637 (8%)
Query: 272 LEREASKENSTKKSSDRQNDIVN--LLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVI 329
+ E E K+ D N LL QP ++G + +QL LNWL + + N I
Sbjct: 153 ITEEEEDEEYLKEEEDGLTGSGNTRLLTQPSCIQG-KMRDYQLAGLNWLIRLYENGINGI 211
Query: 330 LADEMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHG 389
LADEMGLGKT+ + ++ L+ ++ P +V+ P T+GNW+ E + P + V++ G
Sbjct: 212 LADEMGLGKTLQTISLLAYLHEYRGINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLG 271
Query: 390 CAKARAIIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVDEG 449
+ R IR+ A KF++ +TS+EM + + + R W +I+DE
Sbjct: 272 NPEERRHIRE------------DLLVAGKFDICVTSFEMAIKEKTALRRFSWRYIIIDEA 319
Query: 450 HRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEERFN 509
HR+KN S L + S +R+L+TGTPLQNNL E++ LLNFL P F S F+E F
Sbjct: 320 HRIKNENSLLSKTMRLFSTNYRLLITGTPLQNNLHELWALLNFLLPEIFSSAETFDEWFQ 379
Query: 510 ---DLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAMLTK 566
+ E V +L K++ P +LRRLK D + +PPK E ++ V +S +Q +YY+A+L K
Sbjct: 380 ISGENDQQEVVQQLHKVLRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQK 439
Query: 567 NYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKLTL 626
+ L + G ++ +LNI MQLRK CNHPYL G EP G + I + K+ L
Sbjct: 440 D---LEAVNAGGERKRLLNIAMQLRKCCNHPYLFQGAEP--GPPYTTGDHLITNAGKMVL 494
Query: 627 LHSMLKILYKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTYERVDGSVSVTDRQTAIARF 686
L +L L + RVLIFSQMT+LLDILEDYL + Y R+DG+ +R +I +
Sbjct: 495 LDKLLPKLKERDSRVLIFSQMTRLLDILEDYLM--YRGYLYCRIDGNTGGDERDASIEAY 552
Query: 687 NQDKS-RFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSNRLLVY 745
N+ S +FVFLLSTR+ GLGINLATAD VI+YDSD+NP D+QA +RAHRIGQ + V+
Sbjct: 553 NKPGSEKFVFLLSTRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVF 612
Query: 746 RLVVRASVEERILQLAKKKLMLDQLF---------KGKSGSQKEVEDILKWGTEELFNDS 796
R +++EE++++ A KKL LD L K KS ++ E+ ++++G E +F+
Sbjct: 613 RFCTESAIEEKVIERAYKKLALDALVIQQGRLAEQKSKSVNKDELLQMVRYGAEMVFSSK 672
Query: 797 CALNGKDTSENNNSNKDEAVAEVEHKHRKRTGGLGDVYEDKCTDNSSKIMWDENAILKLL 856
+ + + + +EA AE++ K + K T+++ + D++A
Sbjct: 673 DSTITDEDIDRIIAKGEEATAELDAKMK------------KFTEDAIQFKMDDSADFYDF 720
Query: 857 DRSNLQDASTDIAEGDSENDMLGSMKALEWNDEPTEE 893
D N + D + S+N WND P E
Sbjct: 721 DDDNKDENKLDFKKIVSDN----------WNDPPKRE 747
>SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin
dependent regulator of chromatin, subfamily A member 5
(EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog)
(mSnf2h)
Length = 1051
Score = 357 bits (916), Expect = 8e-98
Identities = 212/510 (41%), Positives = 301/510 (58%), Gaps = 38/510 (7%)
Query: 273 EREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILAD 332
E+E +E T+ SS N + P ++ G L +Q+ LNWL + N ILAD
Sbjct: 146 EQEEDEELLTE-SSKATNVCTRFEDSPSYVKWGKLRDYQVRGLNWLISLYENGINGILAD 204
Query: 333 EMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAK 392
EMGLGKT+ + + + + P +VLVP T+ NW++EF W P + V G +
Sbjct: 205 EMGLGKTLQTISLLGYMKHYRNIPGPHMVLVPKSTLHNWMSEFKKWVPTLRSVCLIGDKE 264
Query: 393 ARA-----IIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVD 447
RA ++ EW +V +TSYEM++ + S F+ W L++D
Sbjct: 265 QRAAFVRDVLLPGEW-----------------DVCVTSYEMLIKEKSVFKKFNWRYLVID 307
Query: 448 EGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEER 507
E HR+KN +SKL ++ +R+LLTGTPLQNNL E+++LLNFL P F S F+
Sbjct: 308 EAHRIKNEKSKLSEIVREFKTTNRLLLTGTPLQNNLHELWSLLNFLLPDVFNSADDFDSW 367
Query: 508 F---NDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAML 564
F N L + V+ L ++ P +LRR+K D +++PPK E + V LS +Q E+Y +L
Sbjct: 368 FDTNNCLGDQKLVERLHMVLRPFLLRRIKADVEKSLPPKKEVKIYVGLSKMQREWYTRIL 427
Query: 565 TKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKL 624
K+ IL + GK + + +LNI+MQLRK CNHPYL G EP +H + S K+
Sbjct: 428 MKDIDILNSAGK-MDKMRLLNILMQLRKCCNHPYLFDGAEPGPPYTTDMH--LVTNSGKM 484
Query: 625 TLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYL---NIEFGPKTYERVDGSVSVTDRQT 681
+L +L L ++G RVLIFSQMT++LDILEDY N E Y R+DG +RQ
Sbjct: 485 VVLDKLLPKLKEQGSRVLIFSQMTRVLDILEDYCMWRNYE-----YCRLDGQTPHDERQD 539
Query: 682 AIARFNQDKS-RFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
+I +N+ S +FVF+LSTR+ GLGINLATAD VI+YDSD+NP D+QAM+RAHRIGQ+
Sbjct: 540 SINAYNEPNSTKFVFMLSTRAGGLGINLATADVVILYDSDWNPQVDLQAMDRAHRIGQTK 599
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLDQL 770
+ V+R + +VEERI++ A+ KL LD +
Sbjct: 600 TVRVFRFITDNTVEERIVERAEMKLRLDSI 629
>SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin
dependent regulator of chromatin subfamily A member 5
(EC 3.6.1.-) (SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin A5) (Sucrose
nonfermenting protein 2 homolog) (hSNF2H
Length = 1052
Score = 357 bits (916), Expect = 8e-98
Identities = 212/510 (41%), Positives = 301/510 (58%), Gaps = 38/510 (7%)
Query: 273 EREASKENSTKKSSDRQNDIVNLLEQPKELRGGSLFPHQLEALNWLRKCWYKSRNVILAD 332
E+E +E T+ SS N + P ++ G L +Q+ LNWL + N ILAD
Sbjct: 147 EQEEDEELLTE-SSKATNVCTRFEDSPSYVKWGKLRDYQVRGLNWLISLYENGINGILAD 205
Query: 333 EMGLGKTISACAFISSLYFEFKVSRPCLVLVPLVTMGNWLAEFALWAPDVNVVQYHGCAK 392
EMGLGKT+ + + + + P +VLVP T+ NW++EF W P + V G +
Sbjct: 206 EMGLGKTLQTISLLGYMKHYRNIPGPHMVLVPKSTLHNWMSEFKRWVPTLRSVCLIGDKE 265
Query: 393 ARA-----IIRQYEWHASDPSGLNKKTEAYKFNVLLTSYEMVLADYSHFRGVPWEVLIVD 447
RA ++ EW +V +TSYEM++ + S F+ W L++D
Sbjct: 266 QRAAFVRDVLLPGEW-----------------DVCVTSYEMLIKEKSVFKKFNWRYLVID 308
Query: 448 EGHRLKNSESKLFSLLNSISFQHRVLLTGTPLQNNLGEMYNLLNFLQPASFPSLSAFEER 507
E HR+KN +SKL ++ +R+LLTGTPLQNNL E+++LLNFL P F S F+
Sbjct: 309 EAHRIKNEKSKLSEIVREFKTTNRLLLTGTPLQNNLHELWSLLNFLLPDVFNSADDFDSW 368
Query: 508 F---NDLTSAEKVDELKKLVSPHMLRRLKKDAMQNIPPKTERMVPVELSSIQAEYYRAML 564
F N L + V+ L ++ P +LRR+K D +++PPK E + V LS +Q E+Y +L
Sbjct: 369 FDTNNCLGDQKLVERLHMVLRPFLLRRIKADVEKSLPPKKEVKIYVGLSKMQREWYTRIL 428
Query: 565 TKNYQILRNIGKGIAQQSMLNIVMQLRKVCNHPYLIPGTEPDSGSVEFLHEMRIKASAKL 624
K+ IL + GK + + +LNI+MQLRK CNHPYL G EP +H + S K+
Sbjct: 429 MKDIDILNSAGK-MDKMRLLNILMQLRKCCNHPYLFDGAEPGPPYTTDMH--LVTNSGKM 485
Query: 625 TLLHSMLKILYKEGHRVLIFSQMTKLLDILEDYL---NIEFGPKTYERVDGSVSVTDRQT 681
+L +L L ++G RVLIFSQMT++LDILEDY N E Y R+DG +RQ
Sbjct: 486 VVLDKLLPKLKEQGSRVLIFSQMTRVLDILEDYCMWRNYE-----YCRLDGQTPHDERQD 540
Query: 682 AIARFNQDKS-RFVFLLSTRSCGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSN 740
+I +N+ S +FVF+LSTR+ GLGINLATAD VI+YDSD+NP D+QAM+RAHRIGQ+
Sbjct: 541 SINAYNEPNSTKFVFMLSTRAGGLGINLATADVVILYDSDWNPQVDLQAMDRAHRIGQTK 600
Query: 741 RLLVYRLVVRASVEERILQLAKKKLMLDQL 770
+ V+R + +VEERI++ A+ KL LD +
Sbjct: 601 TVRVFRFITDNTVEERIVERAEMKLRLDSI 630
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 110,047,479
Number of Sequences: 164201
Number of extensions: 4753866
Number of successful extensions: 11786
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 11113
Number of HSP's gapped (non-prelim): 324
length of query: 935
length of database: 59,974,054
effective HSP length: 120
effective length of query: 815
effective length of database: 40,269,934
effective search space: 32819996210
effective search space used: 32819996210
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146651.1


