

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146650.9 - phase: 0 /pseudo
(314 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
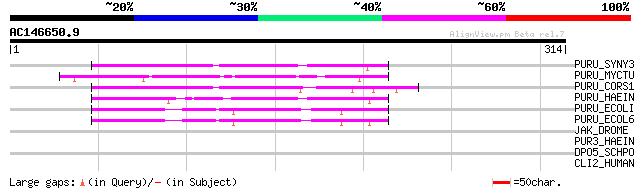
Score E
Sequences producing significant alignments: (bits) Value
PURU_SYNY3 (Q55135) Formyltetrahydrofolate deformylase (EC 3.5.1... 95 3e-19
PURU_MYCTU (Q50453) Formyltetrahydrofolate deformylase (EC 3.5.1... 74 5e-13
PURU_CORS1 (Q46339) Formyltetrahydrofolate deformylase (EC 3.5.1... 68 3e-11
PURU_HAEIN (Q03432) Formyltetrahydrofolate deformylase (EC 3.5.1... 58 3e-08
PURU_ECOLI (P37051) Formyltetrahydrofolate deformylase (EC 3.5.1... 54 5e-07
PURU_ECOL6 (P38480) Formyltetrahydrofolate deformylase (EC 3.5.1... 53 9e-07
JAK_DROME (Q24592) Tyrosine-protein kinase hopscotch (EC 2.7.1.112) 32 2.7
PUR3_HAEIN (P43846) Phosphoribosylglycinamide formyltransferase ... 31 4.6
DPO5_SCHPO (O60094) DNA polymerase V (EC 2.7.7.7) (POL V) 30 6.0
CLI2_HUMAN (O15247) Chloride intracellular channel protein 2 (XA... 30 7.8
>PURU_SYNY3 (Q55135) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 284
Score = 94.7 bits (234), Expect = 3e-19
Identities = 57/172 (33%), Positives = 92/172 (53%), Gaps = 12/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIVA++++ I GNI+ AD + +F +R ++ D + R ++ + +
Sbjct: 12 CPDQPGIVAQIAQFIYQNQGNIIHADQHTDFSSGLFLNRVEWQLDNFRLSRPELLSAWSQ 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L++ AT ++ D ++A+ SKQDHCL+D+L W+ G+L +I +ISNH
Sbjct: 72 LAEQLQATW---QIHFSDQLPRLALWVSKQDHCLLDILWRWRSGELRCEIPLIISNH--- 125
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARYMQ 214
+ ++ GI +HCL T ENK E EL D +VLA+Y+Q
Sbjct: 126 --PDLKSIADQFGIDFHCLPITKENKLAQETAELALLKQYQIDLVVLAKYLQ 175
>PURU_MYCTU (Q50453) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 310
Score = 73.9 bits (180), Expect = 5e-13
Identities = 62/195 (31%), Positives = 94/195 (47%), Gaps = 18/195 (9%)
Query: 29 DLPPLPS---PSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVF--VPQNKHVFY 83
D PP P P G + C D GI+A +S +A G NI++ D P+ F
Sbjct: 16 DYPPPPGGPPPPADIGRLLLRCHDRPGIIAAVSTFLARAGANIISLDQHSTAPEGG-TFL 74
Query: 84 SRSDFVFDPVKWPRKQMEEDF-LKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVD 142
R+ F + +++ DF ++ F A PK ++A++AS +DHCL+D
Sbjct: 75 QRAIFHLPGLTAAVDELQRDFGSTVADKFGIDYRFAE--AAKPK-RVAIMASTEDHCLLD 131
Query: 143 LLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGE---ILE 199
LL + G+L + + VI+N H D HV F G+P+ + T + + E E +
Sbjct: 132 LLWRNRRGELEMSVVMVIAN-HPDLAAHVRPF----GVPFIHIPATRDTRTEAEQRQLQL 186
Query: 200 LVQNTDFLVLARYMQ 214
L N D +VLARYMQ
Sbjct: 187 LSGNVDLVVLARYMQ 201
>PURU_CORS1 (Q46339) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 286
Score = 68.2 bits (165), Expect = 3e-11
Identities = 53/194 (27%), Positives = 89/194 (45%), Gaps = 19/194 (9%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CP+ +GIV ++ + I+ + + + R DF D + +F +
Sbjct: 13 CPEGIGIVHAVTGFLVRHQRTIVELKQYDDMSAGRLFLRVDFAGDSAPDLLDALRSEFSE 72
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH--H 164
++ F+ ++ K K+ ++ SK +HCL DLL G LP+++ V SNH H
Sbjct: 73 VAAKFDMDW---QLRERGQKTKVLIMVSKFEHCLQDLLFRMHSGDLPIEVVGVASNHPDH 129
Query: 165 RDSNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQNT--DFLVLARYMQATG--- 217
R +E +GI +H + + + K E +LEL+ T + +VLARYMQ
Sbjct: 130 RS-------LVEWYGIGFHHIPISKDTKPRAEAALLELIDQTGAELVVLARYMQVLSDHL 182
Query: 218 MT*LTFTMVCCHHS 231
+ LT + HHS
Sbjct: 183 ASELTGKTINIHHS 196
>PURU_HAEIN (Q03432) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 278
Score = 58.2 bits (139), Expect = 3e-08
Identities = 50/174 (28%), Positives = 82/174 (46%), Gaps = 21/174 (12%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDF--VFDPVKWPRKQMEEDF 104
CPD G++AK++ NIL + FV F+ R++ +F+ + ED
Sbjct: 11 CPDDKGLIAKITNICYKHQLNILHNNEFVDFETKHFFMRTELEGIFNEAT-----LLED- 64
Query: 105 LKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHH 164
LK S ++ + +I +L +K+ HCL D+L G L V+I VI NH
Sbjct: 65 LKYSLPEETNCRLIGTQ----RKRIVILVTKEAHCLGDILMKNYYGALDVEIAAVIGNH- 119
Query: 165 RDSNTHVIRFLERHGIPYHCLSTTNENKREGEILELVQ----NTDFLVLARYMQ 214
++ +ER IP+H +S N + E + L + D++VLA+YM+
Sbjct: 120 ----DNLRELVERFNIPFHLVSHENLTRVEHDKLLAEKIDEYTPDYIVLAKYMR 169
>PURU_ECOLI (P37051) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 280
Score = 53.9 bits (128), Expect = 5e-07
Identities = 48/174 (27%), Positives = 80/174 (45%), Gaps = 21/174 (12%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD G++A+++ NI+ + FV F+ R++ + + D
Sbjct: 13 CPDQKGLIARITNICYKHELNIVQNNEFVDHRTGRFFMRTEL---------EGIFNDSTL 63
Query: 107 LSQAFNATRSVVRVPALDP--KYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHH 164
L+ +A V L+P + +I +L +K+ HCL DLL G L V+I VI NH
Sbjct: 64 LADLDSALPEG-SVRELNPAGRRRIVILVTKEAHCLGDLLMKANYGGLDVEIAAVIGNHD 122
Query: 165 RDSNTHVIRFLERHGIPYHCLS----TTNENKREGEILELVQNTDFLVLARYMQ 214
+ +ER IP+ +S T NE+ ++ D++VLA+YM+
Sbjct: 123 T-----LRSLVERFDIPFELVSHEGLTRNEHDQKMADAIDAYQPDYVVLAKYMR 171
>PURU_ECOL6 (P38480) Formyltetrahydrofolate deformylase (EC
3.5.1.10) (Formyl-FH(4) hydrolase)
Length = 280
Score = 53.1 bits (126), Expect = 9e-07
Identities = 46/174 (26%), Positives = 82/174 (46%), Gaps = 21/174 (12%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD G++A+++ NI+ + FV F+ R++ + + D
Sbjct: 13 CPDQKGLIARITNICYKHELNIVQNNEFVDHRTGRFFMRTEL---------EGIFNDSTL 63
Query: 107 LSQAFNATRSVVRVPALDP--KYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHH 164
L+ +A V L+P + +I +L +K+ HCL DLL G L V+I VI NH
Sbjct: 64 LADLDSALPEG-SVRELNPAGRRRIVILVTKEAHCLGDLLMKANYGGLDVEIAAVIGNHD 122
Query: 165 RDSNTHVIRFLERHGIPYHCLS--TTNENKREGEILELVQ--NTDFLVLARYMQ 214
+ +ER IP+ +S + N+ + ++ + + D++VLA+YM+
Sbjct: 123 T-----LRSLVERFDIPFELVSHEGLSRNEHDQKMADAIDAYQPDYVVLAKYMR 171
>JAK_DROME (Q24592) Tyrosine-protein kinase hopscotch (EC 2.7.1.112)
Length = 1177
Score = 31.6 bits (70), Expect = 2.7
Identities = 14/37 (37%), Positives = 23/37 (61%)
Query: 138 HCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRF 174
HCL+DL+HG G ++ +I N+ R SN +V ++
Sbjct: 678 HCLLDLMHGLVRGMHYLEDNKIIHNYIRCSNLYVTKY 714
>PUR3_HAEIN (P43846) Phosphoribosylglycinamide formyltransferase (EC
2.1.2.2) (GART) (GAR transformylase)
(5'-phosphoribosylglycinamide transformylase)
Length = 212
Score = 30.8 bits (68), Expect = 4.6
Identities = 27/92 (29%), Positives = 45/92 (48%), Gaps = 8/92 (8%)
Query: 128 KIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCL-- 185
KIAVL S Q L ++ G +P I CVISN ++ + + ++ IP
Sbjct: 3 KIAVLISGQGTNLQTIIDACHSGDIPAKIACVISN---KADAYGLVRAKQAQIPQAVFLR 59
Query: 186 -STTNENKREGEILELVQN--TDFLVLARYMQ 214
+ +N + + I + +Q+ D +VLA YM+
Sbjct: 60 KNFSNNLEMDDAIGDYLQSLAVDLIVLAGYMK 91
>DPO5_SCHPO (O60094) DNA polymerase V (EC 2.7.7.7) (POL V)
Length = 959
Score = 30.4 bits (67), Expect = 6.0
Identities = 15/60 (25%), Positives = 36/60 (60%), Gaps = 2/60 (3%)
Query: 248 LAQQVTLCPKNLTADQ*LNKWLREFHTEMTCRALCRNQKTLRNNVFPR-LLDPIVNFGYY 306
L V CP+++ +D +W R++ T+ T ++ R++++++ + R LL+ + +GY+
Sbjct: 401 LLSYVKRCPEDIASDTKAVEWRRQWATD-TMLSILRSKRSIKQEPWVRELLEIFIAYGYF 459
>CLI2_HUMAN (O15247) Chloride intracellular channel protein 2
(XAP121)
Length = 243
Score = 30.0 bits (66), Expect = 7.8
Identities = 12/31 (38%), Positives = 19/31 (60%)
Query: 98 KQMEEDFLKLSQAFNATRSVVRVPALDPKYK 128
K+++ DF+K+ + T + R P L PKYK
Sbjct: 77 KELKTDFIKIEEFLEQTLAPPRYPHLSPKYK 107
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.144 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,005,284
Number of Sequences: 164201
Number of extensions: 1308200
Number of successful extensions: 5027
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 5009
Number of HSP's gapped (non-prelim): 10
length of query: 314
length of database: 59,974,054
effective HSP length: 110
effective length of query: 204
effective length of database: 41,911,944
effective search space: 8550036576
effective search space used: 8550036576
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146650.9


