

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.9 - phase: 0 /pseudo
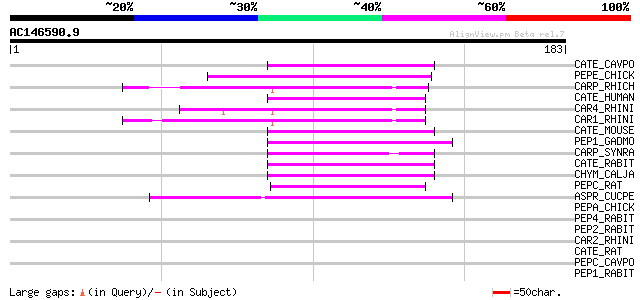
(183 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CATE_CAVPO (P25796) Cathepsin E precursor (EC 3.4.23.34) 47 2e-05
PEPE_CHICK (P16476) Embryonic pepsinogen precursor (EC 3.4.23.-) 47 3e-05
CARP_RHICH (P06026) Rhizopuspepsin precursor (EC 3.4.23.21) 47 3e-05
CATE_HUMAN (P14091) Cathepsin E precursor (EC 3.4.23.34) 44 2e-04
CAR4_RHINI (Q03700) Rhizopuspepsin 4 precursor (EC 3.4.23.21) (A... 44 2e-04
CAR1_RHINI (P10602) Rhizopuspepsin 1 precursor (EC 3.4.23.21) (A... 44 2e-04
CATE_MOUSE (P70269) Cathepsin E precursor (EC 3.4.23.34) 44 2e-04
PEP1_GADMO (P56272) Pepsin IIB (EC 3.4.23.-) 44 3e-04
CARP_SYNRA (P81214) Syncephapepsin precursor (EC 3.4.23.-) 44 3e-04
CATE_RABIT (P43159) Cathepsin E precursor (EC 3.4.23.34) 43 4e-04
CHYM_CALJA (Q9N2D2) Chymosin precursor (EC 3.4.23.4) (Preprorennin) 43 5e-04
PEPC_RAT (P04073) Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C) 42 6e-04
ASPR_CUCPE (O04057) Aspartic proteinase precursor (EC 3.4.23.-) 42 8e-04
PEPA_CHICK (P00793) Pepsin A precursor (EC 3.4.23.1) 42 0.001
PEP4_RABIT (P28713) Pepsin II-4 precursor (EC 3.4.23.1) (Pepsin A) 42 0.001
PEP2_RABIT (P27821) Pepsin II-2/3 precursor (EC 3.4.23.1) (Pepsi... 42 0.001
CAR2_RHINI (P43231) Rhizopuspepsin 2 precursor (EC 3.4.23.21) (A... 41 0.001
CATE_RAT (P16228) Cathepsin E precursor (EC 3.4.23.34) 41 0.002
PEPC_CAVPO (Q64411) Gastricsin precursor (EC 3.4.23.3) (Pepsinog... 40 0.003
PEP1_RABIT (P28712) Pepsin II-1 precursor (EC 3.4.23.1) (Pepsin A) 40 0.003
>CATE_CAVPO (P25796) Cathepsin E precursor (EC 3.4.23.34)
Length = 391
Score = 47.4 bits (111), Expect = 2e-05
Identities = 24/55 (43%), Positives = 30/55 (53%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
EY T S+G+PP I DTGS +W+ C QT P F PS SSTY+ +
Sbjct: 73 EYFGTISIGSPPQNFTVIFDTGSSNLWVPSVYCTSPACQTHPVFHPSLSSTYREV 127
>PEPE_CHICK (P16476) Embryonic pepsinogen precursor (EC 3.4.23.-)
Length = 383
Score = 46.6 bits (109), Expect = 3e-05
Identities = 26/74 (35%), Positives = 36/74 (48%)
Query: 66 HFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQT 125
H + LT + + EY T S+GTPP + DTGS +W+ C Q+
Sbjct: 55 HAFPDVLTVVTEPLLNTLDMEYYGTISIGTPPQDFTVVFDTGSSNLWVPSVSCTSPACQS 114
Query: 126 TPKFKPSKSSTYKN 139
F PS+SSTYK+
Sbjct: 115 HQMFNPSQSSTYKS 128
>CARP_RHICH (P06026) Rhizopuspepsin precursor (EC 3.4.23.21)
Length = 393
Score = 46.6 bits (109), Expect = 3e-05
Identities = 33/114 (28%), Positives = 51/114 (43%), Gaps = 24/114 (21%)
Query: 38 SKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQSTVIPDHG------------ 85
+K+P Y+P+ A+ +I +A Y NT ++PD G
Sbjct: 33 AKNPNYKPS----------AKNAIQKAIAKYNKHKINTSTGGIVPDAGVGTVPMTDYGND 82
Query: 86 -EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYK 138
EY ++GTP K DTGS +W+ C C ++ T K+ P +SSTY+
Sbjct: 83 VEYYGQVTIGTPGKKFNLDFDTGSSDLWIASTLCTNCGSRQT-KYDPKQSSTYQ 135
>CATE_HUMAN (P14091) Cathepsin E precursor (EC 3.4.23.34)
Length = 401
Score = 44.3 bits (103), Expect = 2e-04
Identities = 22/52 (42%), Positives = 30/52 (57%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTY 137
EY T S+G+PP I DTGS +W+ C +T +F+PS+SSTY
Sbjct: 77 EYFGTISIGSPPQNFTVIFDTGSSNLWVPSVYCTSPACKTHSRFQPSQSSTY 128
>CAR4_RHINI (Q03700) Rhizopuspepsin 4 precursor (EC 3.4.23.21)
(Aspartate protease)
Length = 398
Score = 44.3 bits (103), Expect = 2e-04
Identities = 34/99 (34%), Positives = 45/99 (45%), Gaps = 19/99 (19%)
Query: 57 ARRSINRANHFYK---------------TALTNTPQSTVIPDHG---EYLMTYSVGTPPF 98
A+R+I +AN Y +A T + S + D G EY +VGTP
Sbjct: 42 AKRAIEKANAKYARFRSSSSSSSSSSCGSAGTESSGSVPVTDDGNDIEYYGEVTVGTPGI 101
Query: 99 KLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTY 137
KL DTGS +W C C + T K+ PS+SSTY
Sbjct: 102 KLKLDFDTGSSDLWFASTLCTNCGSSQT-KYDPSQSSTY 139
>CAR1_RHINI (P10602) Rhizopuspepsin 1 precursor (EC 3.4.23.21)
(Aspartate protease)
Length = 389
Score = 44.3 bits (103), Expect = 2e-04
Identities = 34/103 (33%), Positives = 46/103 (44%), Gaps = 7/103 (6%)
Query: 38 SKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQSTVIPDHG---EYLMTYSVG 94
+K+ Y+P+ ++ +N A NR T S + D+ EY +VG
Sbjct: 33 AKNNSYKPSA---KNALNKALAKYNRRKVGSGGITTEASGSVPMVDYENDVEYYGEVTVG 89
Query: 95 TPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTY 137
TP KL DTGS +W C C N T K+ P KSSTY
Sbjct: 90 TPGIKLKLDFDTGSSDMWFASTLCSSCSNSHT-KYDPKKSSTY 131
>CATE_MOUSE (P70269) Cathepsin E precursor (EC 3.4.23.34)
Length = 397
Score = 43.9 bits (102), Expect = 2e-04
Identities = 22/55 (40%), Positives = 28/55 (50%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
EY T S+GTPP I DTGS +W+ C + P F PS+S TY +
Sbjct: 78 EYFGTISIGTPPQNFTVIFDTGSSNLWVPSVYCTSPACKAHPVFHPSQSDTYTEV 132
>PEP1_GADMO (P56272) Pepsin IIB (EC 3.4.23.-)
Length = 324
Score = 43.5 bits (101), Expect = 3e-04
Identities = 24/61 (39%), Positives = 29/61 (47%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSSD 145
EY S+GTPP I DTGS +W+ C KFKP +SSTY + D
Sbjct: 13 EYYGVISIGTPPESFKVIFDTGSSNLWVSSSHCSAQACSNHNKFKPRQSSTYVETGKTVD 72
Query: 146 L 146
L
Sbjct: 73 L 73
>CARP_SYNRA (P81214) Syncephapepsin precursor (EC 3.4.23.-)
Length = 395
Score = 43.5 bits (101), Expect = 3e-04
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 3/55 (5%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
EY T SVGTP + DTGS +W C C +++ F P+KSSTYK +
Sbjct: 88 EYYATVSVGTPAQSIKLDFDTGSSDLWFSSTLCTSCGSKS---FDPTKSSTYKKV 139
>CATE_RABIT (P43159) Cathepsin E precursor (EC 3.4.23.34)
Length = 396
Score = 43.1 bits (100), Expect = 4e-04
Identities = 21/55 (38%), Positives = 30/55 (54%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
EY T S+G+PP I DT S +W+ C Q P+F+PS+S+TY +
Sbjct: 77 EYFGTISIGSPPQNFTVIFDTVSSNLWVPSVYCTSPACQMHPQFRPSQSNTYSEV 131
>CHYM_CALJA (Q9N2D2) Chymosin precursor (EC 3.4.23.4) (Preprorennin)
Length = 381
Score = 42.7 bits (99), Expect = 5e-04
Identities = 20/55 (36%), Positives = 30/55 (54%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
+Y +GTPP + + DTGS +W+ C Q +F PSKSST++N+
Sbjct: 73 QYFGKIYIGTPPQEFTVVFDTGSSDLWVPSVYCNSVACQNHHRFDPSKSSTFQNM 127
>PEPC_RAT (P04073) Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C)
Length = 392
Score = 42.4 bits (98), Expect = 6e-04
Identities = 21/51 (41%), Positives = 27/51 (52%)
Query: 87 YLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTY 137
Y S+GTPP + DTGS +W+ C+ T +F PSKSSTY
Sbjct: 76 YFGEISIGTPPQNFLVLFDTGSSNLWVSSVYCQSEACTTHARFNPSKSSTY 126
>ASPR_CUCPE (O04057) Aspartic proteinase precursor (EC 3.4.23.-)
Length = 513
Score = 42.0 bits (97), Expect = 8e-04
Identities = 29/100 (29%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Query: 47 QNKYQHIVNAARRSINRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADT 106
++K I+ AA R N + +++ T+ D +Y ++GTPP K I DT
Sbjct: 51 ESKDAEILKAAFRKYNPKGNLGESSDTDIVALKNYLD-AQYYGEIAIGTPPQKFTVIFDT 109
Query: 107 GSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSSDL 146
GS +W+ CE ++K S+SS+YK S+ +
Sbjct: 110 GSSNLWVLCECLFSVACHFHARYKSSRSSSYKKNGTSASI 149
>PEPA_CHICK (P00793) Pepsin A precursor (EC 3.4.23.1)
Length = 367
Score = 41.6 bits (96), Expect = 0.001
Identities = 26/72 (36%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query: 68 YKTALTNTPQSTVIPDH--GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQT 125
Y LT T + ++ Y T S+GTP I DTGS +W+ CK
Sbjct: 38 YHPVLTATESYEPMTNYMDASYYGTISIGTPQQDFSVIFDTGSSNLWVPSIYCKSSACSN 97
Query: 126 TPKFKPSKSSTY 137
+F PSKSSTY
Sbjct: 98 HKRFDPSKSSTY 109
>PEP4_RABIT (P28713) Pepsin II-4 precursor (EC 3.4.23.1) (Pepsin A)
Length = 387
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/53 (39%), Positives = 26/53 (48%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYK 138
EY T S+GTPP I DTGS +W+ C +F P SSTY+
Sbjct: 74 EYFGTISIGTPPQDFTVIFDTGSSNLWVPSTYCSSLACALHKRFNPEDSSTYQ 126
>PEP2_RABIT (P27821) Pepsin II-2/3 precursor (EC 3.4.23.1) (Pepsin
A)
Length = 387
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/53 (39%), Positives = 26/53 (48%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYK 138
EY T S+GTPP I DTGS +W+ C +F P SSTY+
Sbjct: 74 EYFGTISIGTPPQDFTVIFDTGSSNLWVPSTYCSSLACALHKRFNPEDSSTYQ 126
>CAR2_RHINI (P43231) Rhizopuspepsin 2 precursor (EC 3.4.23.21)
(Aspartate protease)
Length = 391
Score = 41.2 bits (95), Expect = 0.001
Identities = 37/139 (26%), Positives = 64/139 (45%), Gaps = 18/139 (12%)
Query: 7 LILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANH 66
LI +L F+ + A +G + + +K+ Y+P+ NA ++++ + +
Sbjct: 5 LISSCVALAFMALATEAAPSGKKLSI---PLTKNTNYKPSAK------NAIQKALAKYHR 55
Query: 67 FYKTALTNTPQ-----STVIPDHG---EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPC 118
F T+ +N+ S + D+ EY +VGTP L DTGS +W C
Sbjct: 56 FRTTSSSNSTSTEGTGSVPVTDYYNDIEYYGKVTVGTPGVTLKLDFDTGSSDLWFASTLC 115
Query: 119 KECYNQTTPKFKPSKSSTY 137
C + T K+ P++SSTY
Sbjct: 116 TNCGSSQT-KYNPNQSSTY 133
>CATE_RAT (P16228) Cathepsin E precursor (EC 3.4.23.34)
Length = 398
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/55 (38%), Positives = 28/55 (50%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNI 140
EY T S+G+P I DTGS +W+ C + P F PS+SSTY +
Sbjct: 79 EYFGTVSIGSPSQNFTVIFDTGSSNLWVPSVYCTSPACKAHPVFHPSQSSTYMEV 133
>PEPC_CAVPO (Q64411) Gastricsin precursor (EC 3.4.23.3) (Pepsinogen
C)
Length = 394
Score = 40.0 bits (92), Expect = 0.003
Identities = 24/79 (30%), Positives = 35/79 (43%), Gaps = 5/79 (6%)
Query: 64 ANHFYKTALTNTPQSTVIPDHGEYLMT-----YSVGTPPFKLYGIADTGSDIVWLQCEPC 118
A F++ L T +V+ + Y+ S+GTPP + DTGS +W+ C
Sbjct: 51 ARKFFRNRLAKTGDFSVLYEPMSYMDAAYFGQISLGTPPQSFQVLFDTGSSNLWVPSVYC 110
Query: 119 KECYNQTTPKFKPSKSSTY 137
T +F P SSTY
Sbjct: 111 SSLACTTHTRFNPRDSSTY 129
>PEP1_RABIT (P28712) Pepsin II-1 precursor (EC 3.4.23.1) (Pepsin A)
Length = 387
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/53 (37%), Positives = 27/53 (50%)
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYK 138
EY T S+GTPP + I DTGS +W+ C +F P SST++
Sbjct: 74 EYFGTISIGTPPQEFTVIFDTGSSNLWVPSTYCSSLACFLHKRFNPDDSSTFQ 126
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,960,459
Number of Sequences: 164201
Number of extensions: 905640
Number of successful extensions: 2135
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 2064
Number of HSP's gapped (non-prelim): 99
length of query: 183
length of database: 59,974,054
effective HSP length: 103
effective length of query: 80
effective length of database: 43,061,351
effective search space: 3444908080
effective search space used: 3444908080
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146590.9


