

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.2 + phase: 0
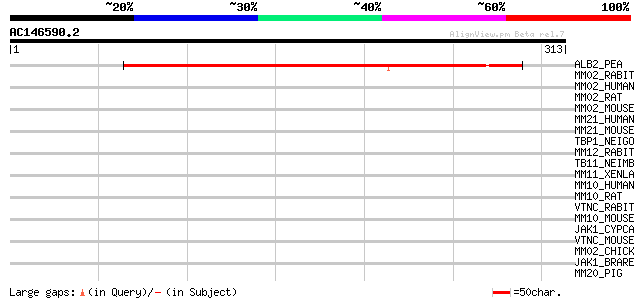
(313 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ALB2_PEA (P08688) Albumin 2 (PA2) 261 2e-69
MM02_RABIT (P50757) 72 kDa type IV collagenase precursor (EC 3.4... 37 0.063
MM02_HUMAN (P08253) 72 kDa type IV collagenase precursor (EC 3.4... 37 0.063
MM02_RAT (P33436) 72 kDa type IV collagenase precursor (EC 3.4.2... 37 0.083
MM02_MOUSE (P33434) 72 kDa type IV collagenase precursor (EC 3.4... 37 0.083
MM21_HUMAN (Q8N119) Matrix metalloproteinase-21 precursor (EC 3.... 35 0.24
MM21_MOUSE (Q8K3F2) Matrix metalloproteinase-21 precursor (EC 3.... 35 0.31
TBP1_NEIGO (Q01996) Transferrin-binding protein 1 precursor 34 0.54
MM12_RABIT (P79227) Macrophage metalloelastase precursor (EC 3.4... 34 0.54
TB11_NEIMB (Q09056) Transferrin-binding protein 1 precursor 33 0.70
MM11_XENLA (Q11005) Stromelysin-3 precursor (EC 3.4.24.-) (Matri... 33 1.2
MM10_HUMAN (P09238) Stromelysin-2 precursor (EC 3.4.24.22) (Matr... 32 1.6
MM10_RAT (P07152) Stromelysin-2 precursor (EC 3.4.24.22) (Matrix... 32 2.0
VTNC_RABIT (P22458) Vitronectin precursor (Serum spreading facto... 32 2.7
MM10_MOUSE (O55123) Stromelysin-2 precursor (EC 3.4.24.22) (Matr... 32 2.7
JAK1_CYPCA (Q09178) Tyrosine-protein kinase Jak1 (EC 2.7.1.112) ... 32 2.7
VTNC_MOUSE (P29788) Vitronectin precursor (Serum spreading facto... 31 3.5
MM02_CHICK (Q90611) 72 kDa type IV collagenase precursor (EC 3.4... 31 3.5
JAK1_BRARE (O12990) Tyrosine-protein kinase Jak1 (EC 2.7.1.112) ... 31 4.5
MM20_PIG (P79287) Matrix metalloproteinase-20 precursor (EC 3.4.... 30 5.9
>ALB2_PEA (P08688) Albumin 2 (PA2)
Length = 231
Score = 261 bits (666), Expect = 2e-69
Identities = 122/227 (53%), Positives = 166/227 (72%), Gaps = 3/227 (1%)
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGI 124
YINAAFR+S NNEAYLF+N++YVL++YA G++ND ++ GP + DG+ SL +T FG +G+
Sbjct: 6 YINAAFRSSQNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRDGFKSLNQTVFGSYGV 65
Query: 125 DCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAF 184
DC+FDTD +A+IF CALIDYAP + DKI+ GP KI DMFPFF+ TVFE G+DAA+
Sbjct: 66 DCSFDTDNDEAFIFYEKFCALIDYAPHSNKDKIILGPKKIADMFPFFEGTVFENGIDAAY 125
Query: 185 RAHSSNEAYLFRGGHYALINYSSKRLIH--IETIRHGFPSLIGTVFENGLEAAFASHGTK 242
R+ E YLF+G YA I+Y + +++ I++IR+GFP T+FE+G +AAFASH T
Sbjct: 126 RSTRGKEVYLFKGDQYARIDYETNSMVNKEIKSIRNGFPCFRNTIFESGTDAAFASHKTN 185
Query: 243 EAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKILPRNN 289
E Y FKG+YYA + PG+TDD ++ G V+ L WPSL+ I+P N
Sbjct: 186 EVYFFKGDYYARVTVTPGATDDQIMDG-VRKTLDYWPSLRGIIPLEN 231
>MM02_RABIT (P50757) 72 kDa type IV collagenase precursor (EC
3.4.24.24) (72 kDa gelatinase) (Matrix
metalloproteinase-2) (MMP-2) (Gelatinase A)
Length = 662
Score = 37.0 bits (84), Expect = 0.063
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 9/79 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGLEAAF 236
+DAAF + + Y+F G + N K++ GFP LI + + L+A
Sbjct: 570 VDAAFNWSKNKKTYIFAGDKFWRYNEVKKKM------DPGFPRLIADAWNAIPDHLDAVV 623
Query: 237 ASHGTKEAYLFKGEYYANI 255
G+ +Y FKG YY +
Sbjct: 624 DLQGSGHSYFFKGTYYLKL 642
Score = 31.6 bits (70), Expect = 2.7
Identities = 38/158 (24%), Positives = 63/158 (39%), Gaps = 21/158 (13%)
Query: 62 IPIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGE 121
+P I+A + A +A F NEY + + ST + PL P + R
Sbjct: 518 LPEKIDAVYEAPQEEKAVFFAGNEYWVYS---ASTLERGYPKPLTSLGLPPDVQR----- 569
Query: 122 HGIDCAFDTDKTQ-AYIFSANLCALIDYAPGTTNDKILSG-PMKITDMFPFFKDTVFERG 179
+D AF+ K + YIF+ + + K+ G P I D + D
Sbjct: 570 --VDAAFNWSKNKKTYIFAGDKF----WRYNEVKKKMDPGFPRLIADAWNAIPDH----- 618
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIR 217
LDA S +Y F+G +Y + S + + + +I+
Sbjct: 619 LDAVVDLQGSGHSYFFKGTYYLKLENQSLKSVKVGSIK 656
>MM02_HUMAN (P08253) 72 kDa type IV collagenase precursor (EC
3.4.24.24) (72 kDa gelatinase) (Matrix
metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE-1)
Length = 660
Score = 37.0 bits (84), Expect = 0.063
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 9/79 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGLEAAF 236
+DAAF + + Y+F G + N K++ GFP LI + + L+A
Sbjct: 568 VDAAFNWSKNKKTYIFAGDKFWRYNEVKKKM------DPGFPKLIADAWNAIPDNLDAVV 621
Query: 237 ASHGTKEAYLFKGEYYANI 255
G +Y FKG YY +
Sbjct: 622 DLQGGGHSYFFKGAYYLKL 640
Score = 30.8 bits (68), Expect = 4.5
Identities = 37/158 (23%), Positives = 61/158 (38%), Gaps = 21/158 (13%)
Query: 62 IPIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGE 121
+P I+A + A +A F NEY + + ST + PL P + R
Sbjct: 516 LPEKIDAVYEAPQEEKAVFFAGNEYWIYS---ASTLERGYPKPLTSLGLPPDVQR----- 567
Query: 122 HGIDCAFDTDKTQ-AYIFSANLCALIDYAPGTTNDKILSG-PMKITDMFPFFKDTVFERG 179
+D AF+ K + YIF+ + + K+ G P I D + D
Sbjct: 568 --VDAAFNWSKNKKTYIFAGDKF----WRYNEVKKKMDPGFPKLIADAWNAIPDN----- 616
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIR 217
LDA +Y F+G +Y + S + + +I+
Sbjct: 617 LDAVVDLQGGGHSYFFKGAYYLKLENQSLKSVKFGSIK 654
>MM02_RAT (P33436) 72 kDa type IV collagenase precursor (EC
3.4.24.24) (72 kDa gelatinase) (Matrix
metalloproteinase-2) (MMP-2) (Gelatinase A)
Length = 662
Score = 36.6 bits (83), Expect = 0.083
Identities = 23/79 (29%), Positives = 34/79 (42%), Gaps = 9/79 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIG---TVFENGLEAAF 236
+DAAF + + Y+F G + N K++ GFP LI + L+A
Sbjct: 570 VDAAFNWSKNKKTYIFAGDKFWRYNEVKKKM------DPGFPKLIADSWNAIPDNLDAVV 623
Query: 237 ASHGTKEAYLFKGEYYANI 255
G +Y FKG YY +
Sbjct: 624 DLQGGGHSYFFKGAYYLKL 642
>MM02_MOUSE (P33434) 72 kDa type IV collagenase precursor (EC
3.4.24.24) (72 kDa gelatinase) (Matrix
metalloproteinase-2) (MMP-2) (Gelatinase A)
Length = 662
Score = 36.6 bits (83), Expect = 0.083
Identities = 23/79 (29%), Positives = 34/79 (42%), Gaps = 9/79 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIG---TVFENGLEAAF 236
+DAAF + + Y+F G + N K++ GFP LI + L+A
Sbjct: 570 VDAAFNWSKNKKTYIFAGDKFWRYNEVKKKM------DPGFPKLIADSWNAIPDNLDAVV 623
Query: 237 ASHGTKEAYLFKGEYYANI 255
G +Y FKG YY +
Sbjct: 624 DLQGGGHSYFFKGAYYLKL 642
>MM21_HUMAN (Q8N119) Matrix metalloproteinase-21 precursor (EC
3.4.24.-) (MMP-21)
Length = 569
Score = 35.0 bits (79), Expect = 0.24
Identities = 33/131 (25%), Positives = 57/131 (43%), Gaps = 16/131 (12%)
Query: 76 NEAYLFMNNEYVLINYARGSTNDYIINGPLY---ICDGYPSLARTPFGEHGIDCAF-DTD 131
+E Y F N+Y + + G Y I +G+P + +D AF D
Sbjct: 406 DERYFFQGNQYWRYDSDKDQALTEDEQGKSYPKLISEGFPGIPSP------LDTAFYDRR 459
Query: 132 KTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFF--KDTVFERGLDAAFRAHSS 189
+ Y F +L D N + S P +IT++FP ++ F R +D+A+ +++
Sbjct: 460 QKLIYFFKESLVFAFDV---NRNRVLNSYPKRITEVFPAVIPQNHPF-RNIDSAYYSYAY 515
Query: 190 NEAYLFRGGHY 200
N + F+G Y
Sbjct: 516 NSIFFFKGNAY 526
>MM21_MOUSE (Q8K3F2) Matrix metalloproteinase-21 precursor (EC
3.4.24.-) (MMP-21)
Length = 568
Score = 34.7 bits (78), Expect = 0.31
Identities = 31/130 (23%), Positives = 53/130 (39%), Gaps = 14/130 (10%)
Query: 76 NEAYLFMNNEYVLINYARGSTNDYIING---PLYICDGYPSLARTPFGEHGIDCAF-DTD 131
+E Y F ++Y + + G P I +G+P + +D AF D
Sbjct: 405 DERYFFKGSQYWRYDSENDQAHTEDEQGRSYPKLISEGFPGIPSP------LDTAFYDRR 458
Query: 132 KTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFE-RGLDAAFRAHSSN 190
+ Y F +L D N + S P K++ +FP R LD+A+ +++ N
Sbjct: 459 QQLIYFFKESLVFAFDV---NRNQVLNSYPKKMSQVFPAIMPQNHPFRNLDSAYYSYAHN 515
Query: 191 EAYLFRGGHY 200
+ F+G Y
Sbjct: 516 SIFFFKGNSY 525
>TBP1_NEIGO (Q01996) Transferrin-binding protein 1 precursor
Length = 915
Score = 33.9 bits (76), Expect = 0.54
Identities = 14/46 (30%), Positives = 27/46 (58%)
Query: 15 IDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVE 60
ID NG+ +LPE Y+ ++V+ RD+ D +++ H + A++
Sbjct: 742 IDWNGVWDKLPEGWYSTFAYNRVRVRDIKKRADRTDIQSHLFDAIQ 787
>MM12_RABIT (P79227) Macrophage metalloelastase precursor (EC
3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12)
Length = 464
Score = 33.9 bits (76), Expect = 0.54
Identities = 24/104 (23%), Positives = 50/104 (48%), Gaps = 10/104 (9%)
Query: 179 GLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRH-GFPSLIGTVFENGLEAAFA 237
G++AA+ ++ +LF+G + LI++ + + ++I GFP F ++AA
Sbjct: 326 GIEAAYEIGDRHQVFLFKGDKFWLISHLRLQPNYPKSIHSLGFPD-----FVKKIDAAVF 380
Query: 238 SHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSL 281
+ ++ Y F N+Y+ + + G KLI ++P +
Sbjct: 381 NPSLRKTYFF----VDNLYWRYDERREVMDAGYPKLITKHFPGI 420
>TB11_NEIMB (Q09056) Transferrin-binding protein 1 precursor
Length = 911
Score = 33.5 bits (75), Expect = 0.70
Identities = 14/46 (30%), Positives = 26/46 (56%)
Query: 15 IDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVE 60
ID NG+ +LPE Y+ ++V RD+ D +++ H + A++
Sbjct: 738 IDWNGVWDKLPEGWYSTFAYNRVHVRDIKKRADRTDIQSHLFDAIQ 783
>MM11_XENLA (Q11005) Stromelysin-3 precursor (EC 3.4.24.-) (Matrix
metalloproteinase-11) (MMP-11) (ST3)
Length = 477
Score = 32.7 bits (73), Expect = 1.2
Identities = 39/148 (26%), Positives = 59/148 (39%), Gaps = 21/148 (14%)
Query: 109 DGYPSLARTPFG--EHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITD 166
+GYP+LA + +D AF+ + F + + D SGP ITD
Sbjct: 311 NGYPALASRHWRGIPDTVDAAFEDSVGNIWFFYGSQFWVFD------GKLQASGPFPITD 364
Query: 167 MFPFFKDTVFERGLDAAF--RAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLI 224
+ + + AAF + + YLFRGG Y N ++R+ + R G +
Sbjct: 365 I------GISVTQIQAAFVWGTEKNKKTYLFRGGEYWRFNPETRRVESRHSRRIGDWRGV 418
Query: 225 GTVFENGLEAAFASHGTKEAYLFKGEYY 252
G++AAF AY KG Y
Sbjct: 419 ----PKGIDAAFQDE-QGYAYFVKGRQY 441
>MM10_HUMAN (P09238) Stromelysin-2 precursor (EC 3.4.24.22) (Matrix
metalloproteinase-10) (MMP-10) (Transin-2) (SL-2)
Length = 476
Score = 32.3 bits (72), Expect = 1.6
Identities = 43/171 (25%), Positives = 68/171 (39%), Gaps = 28/171 (16%)
Query: 48 NEEVELHSYGAV-ENIPIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLY 106
N E E H A ++P Y++AA+ ++ + ++F NE+ I RG+
Sbjct: 320 NPEPEFHLISAFWPSLPSYLDAAYEVNSRDTVFIFKGNEFWAI---RGNE---------- 366
Query: 107 ICDGYPSLART---PFGEHGIDCAF-DTDKTQAYIFSANLCALIDYAPGTTNDKILSGPM 162
+ GYP T P ID A D +K + Y F+A+ D + P
Sbjct: 367 VQAGYPRGIHTLGFPPTIRKIDAAVSDKEKKKTYFFAADKYWRFDENSQSMEQGF---PR 423
Query: 163 KITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHI 213
I D FP + V DA +A Y F G + +++ + HI
Sbjct: 424 LIADDFPGVEPKV-----DAVLQAFGF--FYFFSGSSQFEFDPNARMVTHI 467
>MM10_RAT (P07152) Stromelysin-2 precursor (EC 3.4.24.22) (Matrix
metalloproteinase-10) (MMP-10) (Transin-2) (SL-2)
(Transformation-associated protein 34A)
Length = 476
Score = 32.0 bits (71), Expect = 2.0
Identities = 24/105 (22%), Positives = 48/105 (44%), Gaps = 10/105 (9%)
Query: 179 GLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRH-GFPSLIGTVFENGLEAAFA 237
GLDAA+ A++ + +F+G + + + + + + I GFP + ++AA
Sbjct: 338 GLDAAYEANNKDRVLIFKGSQFWAVRGNEVQAGYPKRIHTLGFPPTV-----KKIDAAVF 392
Query: 238 SHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLK 282
K+ Y F G+ Y+ T + G +LI ++P ++
Sbjct: 393 EKEKKKTYFFVGDK----YWRFDETRQLMDKGFPRLITDDFPGIE 433
>VTNC_RABIT (P22458) Vitronectin precursor (Serum spreading factor)
(S-protein) (Glycoprotein 66)
Length = 475
Score = 31.6 bits (70), Expect = 2.7
Identities = 24/108 (22%), Positives = 41/108 (37%), Gaps = 1/108 (0%)
Query: 177 ERGLDAAF-RAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFENGLEAA 235
E +DAAF R + + YLF+G Y + I GF + V A
Sbjct: 203 EGPIDAAFTRINCQGKTYLFKGSQYWRFEDGILDPDYPRNISEGFSGIPDNVDAAFALPA 262
Query: 236 FASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKK 283
+ G + Y FKG+ Y + + + G + + ++ L +
Sbjct: 263 HSYSGRERVYFFKGDKYWEYQFQQQPSQEECEGSSLSAVFEHFAMLHR 310
>MM10_MOUSE (O55123) Stromelysin-2 precursor (EC 3.4.24.22) (Matrix
metalloproteinase-10) (MMP-10) (Transin-2) (SL-2)
Length = 476
Score = 31.6 bits (70), Expect = 2.7
Identities = 19/74 (25%), Positives = 36/74 (47%), Gaps = 6/74 (8%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRH-GFPSLIGTVFENGLEAAFAS 238
LDAA+ AH+++ +F+G + + + + + + I GFP + ++AA
Sbjct: 339 LDAAYEAHNTDSVLIFKGSQFWAVRGNEVQAGYPKGIHTLGFPPTV-----KKIDAAVFE 393
Query: 239 HGTKEAYLFKGEYY 252
K+ Y F G+ Y
Sbjct: 394 KEKKKTYFFVGDKY 407
>JAK1_CYPCA (Q09178) Tyrosine-protein kinase Jak1 (EC 2.7.1.112)
(Janus kinase 1) (Jak-1)
Length = 1156
Score = 31.6 bits (70), Expect = 2.7
Identities = 22/58 (37%), Positives = 32/58 (54%), Gaps = 5/58 (8%)
Query: 242 KEAYLFKGEYYANIYYAPGSTDDYLIGGR-VKLILSNWP--SLKKILPRNNGRLDLHT 296
+E ++ + Y+ NI G + GGR +KLI+ P SLK+ LPRN +DL T
Sbjct: 924 REIHILRELYHENIVKYKGIWHEE--GGRSIKLIMEFLPAGSLKEYLPRNKAHIDLKT 979
>VTNC_MOUSE (P29788) Vitronectin precursor (Serum spreading factor)
(S-protein)
Length = 478
Score = 31.2 bits (69), Expect = 3.5
Identities = 27/113 (23%), Positives = 48/113 (41%), Gaps = 11/113 (9%)
Query: 177 ERGLDAAF-RAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFENGLEAA 235
E +DAAF R + + YLF+G Y + I GF + + ++AA
Sbjct: 202 EGPIDAAFTRINCQGKTYLFKGSQYWRFEDGVLDPGYPRNISEGFSGI-----PDNVDAA 256
Query: 236 FA--SH---GTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKK 283
FA +H G + Y FKG+ Y + + + G + + ++ L++
Sbjct: 257 FALPAHRYSGRERVYFFKGKQYWEYEFQQQPSQEECEGSSLSAVFEHFALLQR 309
>MM02_CHICK (Q90611) 72 kDa type IV collagenase precursor (EC
3.4.24.24) (72 kDa gelatinase) (Matrix
metalloproteinase-2) (MMP-2) (Gelatinase A)
Length = 663
Score = 31.2 bits (69), Expect = 3.5
Identities = 35/150 (23%), Positives = 61/150 (40%), Gaps = 17/150 (11%)
Query: 103 GPLYICDGYPSLARTPFGEHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPM 162
GPL + +P L ID +++ + + +F A +Y T ++ P
Sbjct: 508 GPLLVATFWPDLPEK------IDAVYESPQDEKAVFFAGN----EYWVYTASNLDRGYPK 557
Query: 163 KITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPS 222
K+T + D + +DAAF + + Y+F G Y N K++ + T + S
Sbjct: 558 KLTSL-GLPPDV---QRIDAAFNWGRNKKTYIFSGDRYWKYNEEKKKM-ELATPKFIADS 612
Query: 223 LIGTVFENGLEAAFASHGTKEAYLFKGEYY 252
G + L+A + Y FK +YY
Sbjct: 613 WNGV--PDNLDAVLGLTDSGYTYFFKDQYY 640
>JAK1_BRARE (O12990) Tyrosine-protein kinase Jak1 (EC 2.7.1.112)
(Janus kinase 1) (Jak-1)
Length = 1153
Score = 30.8 bits (68), Expect = 4.5
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 5/58 (8%)
Query: 242 KEAYLFKGEYYANIYYAPGSTDDYLIGGR-VKLILSNWP--SLKKILPRNNGRLDLHT 296
+E ++ + Y+ NI G ++ GGR +KLI+ P SLK+ LPRN ++L T
Sbjct: 921 REIHILRELYHENIVKYKGICNEE--GGRSIKLIMEFLPAGSLKEYLPRNKAHINLKT 976
>MM20_PIG (P79287) Matrix metalloproteinase-20 precursor (EC
3.4.24.-) (MMP-20) (Enamel metalloproteinase)
(Enamelysin)
Length = 483
Score = 30.4 bits (67), Expect = 5.9
Identities = 28/100 (28%), Positives = 40/100 (40%), Gaps = 13/100 (13%)
Query: 157 ILSG--PMKITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIE 214
++SG P IT FP V DAA+ AY F+G HY + +
Sbjct: 327 LMSGIRPSTITSSFPQLMSNV-----DAAYEVADRGMAYFFKGPHYWITRGFQMQGPPRT 381
Query: 215 TIRHGFPSLIGTVFENGLEAAFASHGTKEAYLFKG-EYYA 253
GFP + ++AA T++ F G EYY+
Sbjct: 382 IYDFGFPRYV-----QRIDAAVHLKDTQKTLFFVGDEYYS 416
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,068,863
Number of Sequences: 164201
Number of extensions: 1841120
Number of successful extensions: 3650
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 3613
Number of HSP's gapped (non-prelim): 36
length of query: 313
length of database: 59,974,054
effective HSP length: 110
effective length of query: 203
effective length of database: 41,911,944
effective search space: 8508124632
effective search space used: 8508124632
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146590.2


