

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.6 - phase: 0 /pseudo
(670 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
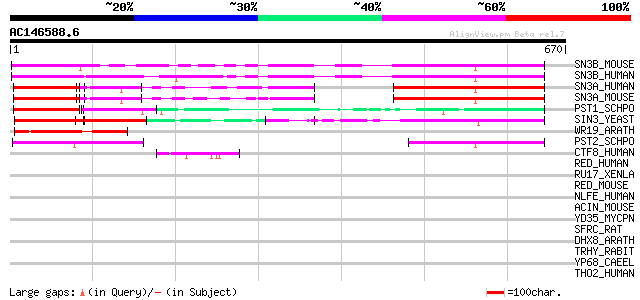
Score E
Sequences producing significant alignments: (bits) Value
SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b 246 2e-64
SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b 238 3e-62
SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a 151 5e-36
SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a 150 1e-35
PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3 ... 150 1e-35
SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3 117 1e-25
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 104 9e-22
PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3 ... 99 3e-20
CTF8_HUMAN (Q96D72) Protein C20orf158 45 6e-04
RED_HUMAN (Q13123) Red protein (RER protein) (IK factor) (Cytoki... 44 0.002
RU17_XENLA (P09406) U1 small nuclear ribonucleoprotein 70 kDa (U... 43 0.002
RED_MOUSE (Q9Z1M8) Red protein (RER protein) 43 0.002
NLFE_HUMAN (P18615) Negative elongation factor E (NELF-E) (RD pr... 42 0.005
ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in ... 42 0.005
YD35_MYCPN (P75443) Hypothetical protein MPN335 (F10_orf741) 41 0.012
SFRC_RAT (Q9JKL7) Splicing factor, arginine/serine-rich 12 (Seri... 41 0.012
DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-depend... 41 0.012
TRHY_RABIT (P37709) Trichohyalin 40 0.015
YP68_CAEEL (Q09217) Hypothetical protein B0495.8 in chromosome II 40 0.020
THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2) 40 0.020
>SN3B_MOUSE (Q62141) Paired amphipathic helix protein Sin3b
Length = 954
Score = 246 bits (627), Expect = 2e-64
Identities = 202/684 (29%), Positives = 297/684 (42%), Gaps = 169/684 (24%)
Query: 3 GGQKLTTN--DALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKG 60
G +KL + DAL+YL V+ F +D Y+ FLE+MK+FK+Q IDT GVI RV +LF
Sbjct: 26 GHEKLPVHVEDALTYLDQVKIRFGSDPATYNGFLEIMKEFKSQSIDTPGVIRRVSQLFHE 85
Query: 61 HKDLILGFNTFLPKGYAITLPSDD----------------------------------EQ 86
H DLI+GFN FLP GY I +P + +
Sbjct: 86 HPDLIVGFNAFLPLGYRIDIPKNGKLNIQSPLSSQDNSHSHGDCGEDFKQMSYKEDRGQV 145
Query: 87 PLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQD 146
PL+ VEF AI++V KIK RF LD +YR L+ + +E
Sbjct: 146 PLESDSVEFNNAISYVNKIKTRF----------LDHPEIYRSFLEILHTYQKE------- 188
Query: 147 HGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMHVDKREKTTTLHADRD 206
+ T P + F N L R + ++ Q + A R
Sbjct: 189 -----QLHTKGRPFRGMSEEEVFTEVAN-LFRGQEDLLSEFGQFLPE---------AKRS 233
Query: 207 LSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGNLERLPHKKK 266
L + E++ G Q+ E+ + ++R R R P KKK
Sbjct: 234 LFTGNGSCEMNSG-------QKNEEKSLEHNKKRSRPSLLRP----------VSAPAKKK 276
Query: 267 SVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDDY 326
R T D + + T ++ S F +KV+ LK+ + Y
Sbjct: 277 MKLRGTK----------DLSIAAVGKYGTLQEFS-----------FFDKVRRVLKSQEVY 315
Query: 327 QEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNKKS 386
+ FL+C+ ++++E+++ ELL LV LGK+ ++ F F+
Sbjct: 316 ENFLRCIALFNQELVSGSELLQLVSPFLGKFPELFAQFKSFL------------------ 357
Query: 387 LWNEGHGQKPLKVEEKDRDRGRDDGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTS 446
G K L DR D +E D +R S+
Sbjct: 358 ------GVKELSFAPPMSDRSGDGISREIDYASCKRIGSS-------------------- 391
Query: 447 M*ESL*MNWTFLTVNNVLPATVSYPKIQ-KTELGAKVLNDHWVSVTSGSEDYSFKHMRKN 505
LP T PK +T + +VLND WVS S SED +F +K
Sbjct: 392 --------------YRALPKTYQQPKCSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKT 437
Query: 506 QYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSA---- 561
YEE L RCED+RFELD++LE+ AT + +E + +K++ +R+++ L
Sbjct: 438 PYEEQLHRCEDERFELDVVLETNLATIRVLESVQKKLSRMAPEDQEKLRLDDCLGGTSEV 497
Query: 562 LNLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAK 621
+ R I R+YGD +V+E LK+N + A+PV+L RLK K+EEW ++ FNK+W E Y K
Sbjct: 498 IQRRAIHRIYGDKAPEVIESLKRNPATAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEK 557
Query: 622 NYHKSLDHRSFYFKQQDTKSLSTK 645
Y KSLDH++ FKQ DTK+L +K
Sbjct: 558 AYLKSLDHQAVNFKQNDTKALRSK 581
>SN3B_HUMAN (O75182) Paired amphipathic helix protein Sin3b
Length = 1130
Score = 238 bits (608), Expect = 3e-62
Identities = 191/658 (29%), Positives = 286/658 (43%), Gaps = 115/658 (17%)
Query: 3 GGQKLTTN--DALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKG 60
G +KL + DAL+YL V+ F +D Y+ FLE+MK+FK+Q IDT GVI RV +LF
Sbjct: 33 GHEKLPVHVEDALTYLDQVKIRFGSDPATYNGFLEIMKEFKSQSIDTPGVIRRVSQLFHE 92
Query: 61 HKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFL 120
H DLI+GFN FLP GY I +P + + +Q P+ +E + G F+ +
Sbjct: 93 HPDLIVGFNAFLPLGYRIDIPKNGKLNIQS-PLTSQENSHNHGDGAEDFKQQVPYKEDKP 151
Query: 121 DI-LNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRD 179
+ L E + ++ F DH ++ F L
Sbjct: 152 QVPLESDSVEFNNAISYVNKIKTRFLDHPEIYRSFLEILHTYQ----------------- 194
Query: 180 RSSAMTTGRQMHVDKREKTTT-----LHADRDLSVDHPD--PELDRGVMRTDKEQRRRER 232
+ T GR E+ T DL + PE R + +
Sbjct: 195 KEQLNTRGRPFRGMSEEEVFTEVANLFRGQEDLLSEFGQFLPEAKRSLFTGNGPCEMHSV 254
Query: 233 EKDRREERDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPN 292
+K+ ++ R+R + P KKK R T D + +
Sbjct: 255 QKNEHDKTPEHSRKRSRPSLLRPVSA---PAKKKMKLRGTK----------DLSIAAVGK 301
Query: 293 SSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGD 352
T ++ S F +KV+ LK+ + Y+ FL+C+ ++++E+++ ELL LV
Sbjct: 302 YGTLQEFS-----------FFDKVRRVLKSQEVYENFLRCIALFNQELVSGSELLQLVSP 350
Query: 353 LLGKYADIMEGFDDFVTQCEKNEGFLAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRDDGV 412
LGK+ ++ F F+ G K L DR D
Sbjct: 351 FLGKFPELFAQFKSFL------------------------GVKELSFAPPMSDRSGDGIS 386
Query: 413 KERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPK 472
+E D +R S+ LP T PK
Sbjct: 387 REIDYASCKRIGSS----------------------------------YRALPKTYQQPK 412
Query: 473 IQ-KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINAT 531
+T + +VLND WVS S SED +F +K YEE L RCED+RFELD++LE+ AT
Sbjct: 413 CSGRTAICKEVLNDTWVSFPSWSEDSTFVSSKKTPYEEQLHRCEDERFELDVVLETNLAT 472
Query: 532 SKKVEEIIEKVNDDIIPGDIPIRIEEHLSA----LNLRCIERLYGDHGLDVMEVLKKNAS 587
+ +E + +K++ R+++ L + R I R+YGD +++E LKKN
Sbjct: 473 IRVLESVQKKLSRMAPEDQEKFRLDDSLGGTSEVIQRRAIYRIYGDKAPEIIESLKKNPV 532
Query: 588 LALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
A+PV+L RLK K+EEW ++ FNK+W E Y K Y KSLDH++ FKQ DTK+L +K
Sbjct: 533 TAVPVVLKRLKAKEEEWREAQQGFNKIWREQYEKAYLKSLDHQAVNFKQNDTKALRSK 590
>SN3A_HUMAN (Q96ST3) Paired amphipathic helix protein Sin3a
Length = 1273
Score = 151 bits (382), Expect = 5e-36
Identities = 80/187 (42%), Positives = 116/187 (61%), Gaps = 5/187 (2%)
Query: 464 LPATVSYPKIQ-KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELD 522
LP + PK +T L +VLND WVS S SED +F +K QYEE ++RCED+RFELD
Sbjct: 561 LPKSYQQPKCTGRTPLCKEVLNDTWVSFPSWSEDSTFVSSKKTQYEEHIYRCEDERFELD 620
Query: 523 MLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSA----LNLRCIERLYGDHGLDV 578
++LE+ AT + +E I +K++ R++ L ++ + ++R+Y D D+
Sbjct: 621 VVLETNLATIRVLEAIQKKLSRLSAEEQAKFRLDNTLGGTSEVIHRKALQRIYADKAADI 680
Query: 579 MEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQD 638
++ L+KN S+A+P++L RLK K+EEW + FNKVW E K Y KSLDH+ FKQ D
Sbjct: 681 IDGLRKNPSIAVPIVLKRLKMKEEEWREAQRGFNKVWREQNEKYYLKSLDHQGINFKQND 740
Query: 639 TKSLSTK 645
TK L +K
Sbjct: 741 TKVLRSK 747
Score = 101 bits (251), Expect = 7e-21
Identities = 82/293 (27%), Positives = 128/293 (42%), Gaps = 85/293 (29%)
Query: 91 KPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPI-------------TAVY 137
+PVEF AIN+V KIKNRFQG +YK FL+IL+ Y+KE + VY
Sbjct: 300 QPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEVY 359
Query: 138 QEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMHVDKREK 197
+V+ LF++ DLL EF FLPD A + +L +++A EK
Sbjct: 360 AQVARLFKNQEDLLSEFGQFLPD-----------ANSSVLLSKTTA------------EK 396
Query: 198 TTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGN 257
++ D +V P + + R N
Sbjct: 397 VDSVRNDHGGTVKKP------------------------------QLNNKPQRPSQNGCQ 426
Query: 258 LERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDK--SSLKSLCSPVLAFLEK 315
+ R P GT P+ + L+L +S K +SL F +K
Sbjct: 427 IRRHP-----------TGTTPPVKKKPKLLNLKDSSMADASKHGGGTESL------FFDK 469
Query: 316 VKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
V++ L++ + Y+ FL+CL I+++E+I+R EL+ LV LGK+ ++ F +F+
Sbjct: 470 VRKALRSAEAYENFLRCLVIFNQEVISRAELVQLVSPFLGKFPELFNWFKNFL 522
Score = 97.8 bits (242), Expect = 8e-20
Identities = 45/80 (56%), Positives = 60/80 (74%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
Q+L DALSYL V+ F + + Y++FL++MK+FK+Q IDT GVI RV +LFKGH DL
Sbjct: 119 QRLKVEDALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDL 178
Query: 65 ILGFNTFLPKGYAITLPSDD 84
I+GFNTFLP GY I + ++D
Sbjct: 179 IMGFNTFLPPGYKIEVQTND 198
Score = 49.7 bits (117), Expect = 3e-05
Identities = 26/79 (32%), Positives = 43/79 (53%), Gaps = 2/79 (2%)
Query: 81 PSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEV 140
P +Q Q+ VE +A++++ ++K +F +VY FLDI+ ++ + V V
Sbjct: 111 PVQGQQQFQRLKVE--DALSYLDQVKLQFGSQPQVYNDFLDIMKEFKSQSIDTPGVISRV 168
Query: 141 SALFQDHGDLLEEFTHFLP 159
S LF+ H DL+ F FLP
Sbjct: 169 SQLFKGHPDLIMGFNTFLP 187
Score = 39.3 bits (90), Expect = 0.034
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 13/95 (13%)
Query: 1 MTGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQR-------------IDT 47
+ Q + N A++Y+ ++ FQ + Y FLE++ ++ ++ +
Sbjct: 296 LQNNQPVEFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTE 355
Query: 48 AGVIERVKELFKGHKDLILGFNTFLPKGYAITLPS 82
V +V LFK +DL+ F FLP + L S
Sbjct: 356 QEVYAQVARLFKNQEDLLSEFGQFLPDANSSVLLS 390
Score = 35.4 bits (80), Expect = 0.49
Identities = 34/148 (22%), Positives = 58/148 (38%), Gaps = 28/148 (18%)
Query: 312 FLEKVKEKLKN-PDDYQEFLKCLHIYSRE-------------IITRQELLALVGDLLGKY 357
++ K+K + + PD Y+ FL+ LH Y +E +T QE+ A V L
Sbjct: 310 YVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEVYAQVARLFKNQ 369
Query: 358 ADIMEGFDDFV-----------TQCEKNEGFL---AGVMNKKSLWNEGHGQKPLKVEEKD 403
D++ F F+ T EK + G + K L N+ + +
Sbjct: 370 EDLLSEFGQFLPDANSSVLLSKTTAEKVDSVRNDHGGTVKKPQLNNKPQRPSQNGCQIRR 429
Query: 404 RDRGRDDGVKERDRELRERDKSTGISNK 431
G VK++ + L +D S ++K
Sbjct: 430 HPTGTTPPVKKKPKLLNLKDSSMADASK 457
>SN3A_MOUSE (Q60520) Paired amphipathic helix protein Sin3a
Length = 1282
Score = 150 bits (379), Expect = 1e-35
Identities = 79/187 (42%), Positives = 116/187 (61%), Gaps = 5/187 (2%)
Query: 464 LPATVSYPKIQ-KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELD 522
LP + PK +T L +VLND WVS S SED +F +K QYEE ++RCED+RFELD
Sbjct: 562 LPKSYQQPKCTGRTPLCKEVLNDTWVSFPSWSEDSTFVSSKKTQYEEHIYRCEDERFELD 621
Query: 523 MLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSA----LNLRCIERLYGDHGLDV 578
++LE+ AT + +E I +K++ R++ L ++ + ++R+Y D D+
Sbjct: 622 VVLETNLATIRVLEAIQKKLSRLSAEEQAKFRLDNTLGGTSEVIHRKALQRIYADKAADI 681
Query: 579 MEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQD 638
++ L+KN S+A+P++L RLK K+EEW + FNKVW + K Y KSLDH+ FKQ D
Sbjct: 682 IDGLRKNPSIAVPIVLKRLKMKEEEWREAQRGFNKVWRDENEKYYLKSLDHQGINFKQND 741
Query: 639 TKSLSTK 645
TK L +K
Sbjct: 742 TKVLRSK 748
Score = 95.9 bits (237), Expect = 3e-19
Identities = 44/80 (55%), Positives = 60/80 (75%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
Q+L DALSYL V+ F + + +++FL++MK+FK+Q IDT GVI RV +LFKGH DL
Sbjct: 119 QRLKVEDALSYLDQVKLQFGSQPQVHNDFLDIMKEFKSQSIDTPGVISRVSQLFKGHPDL 178
Query: 65 ILGFNTFLPKGYAITLPSDD 84
I+GFNTFLP GY I + ++D
Sbjct: 179 IMGFNTFLPPGYKIEVQTND 198
Score = 95.5 bits (236), Expect = 4e-19
Identities = 84/291 (28%), Positives = 129/291 (43%), Gaps = 80/291 (27%)
Query: 91 KPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPI-------------TAVY 137
+PV F AIN+V KIKNRFQG +YK FL+IL+ Y+KE + VY
Sbjct: 300 QPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTEQEVY 359
Query: 138 QEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMHVDKREK 197
+V+ LF++ DLL EF FLPD A + +L +++A EK
Sbjct: 360 AQVARLFKNQEDLLSEFGQFLPD-----------ANSSVLLSKTTA------------EK 396
Query: 198 TTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGN 257
++ D +V P +K QR + R + G
Sbjct: 397 VDSVRNDHGGTVKKP--------QLNNKPQRPSQNGCQIRR---------------HSGT 433
Query: 258 LERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVK 317
P KKK KL L SS + +S + + L F +KV+
Sbjct: 434 GATPPVKKK------------------PKLMSLKESSMAD--ASKHGVGTESLFF-DKVR 472
Query: 318 EKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
+ L++ + Y+ FL+CL I+++E+I+R EL+ LV LGK+ ++ F +F+
Sbjct: 473 KALRSAEAYENFLRCLVIFNQEVISRAELVQLVSPFLGKFPELFNWFKNFL 523
Score = 47.8 bits (112), Expect = 1e-04
Identities = 25/79 (31%), Positives = 43/79 (53%), Gaps = 2/79 (2%)
Query: 81 PSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEV 140
P +Q Q+ VE +A++++ ++K +F +V+ FLDI+ ++ + V V
Sbjct: 111 PVQGQQQFQRLKVE--DALSYLDQVKLQFGSQPQVHNDFLDIMKEFKSQSIDTPGVISRV 168
Query: 141 SALFQDHGDLLEEFTHFLP 159
S LF+ H DL+ F FLP
Sbjct: 169 SQLFKGHPDLIMGFNTFLP 187
Score = 38.9 bits (89), Expect = 0.044
Identities = 23/95 (24%), Positives = 42/95 (44%), Gaps = 13/95 (13%)
Query: 1 MTGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQR-------------IDT 47
+ Q + N A++Y+ ++ FQ + Y FLE++ ++ ++ +
Sbjct: 296 LQNNQPVHFNHAINYVNKIKNRFQGQPDIYKAFLEILHTYQKEQRNAKEAGGNYTPALTE 355
Query: 48 AGVIERVKELFKGHKDLILGFNTFLPKGYAITLPS 82
V +V LFK +DL+ F FLP + L S
Sbjct: 356 QEVYAQVARLFKNQEDLLSEFGQFLPDANSSVLLS 390
>PST1_SCHPO (Q09750) Paired amphipathic helix protein pst1 (SIN3
homolog 1)
Length = 1522
Score = 150 bits (378), Expect = 1e-35
Identities = 149/576 (25%), Positives = 222/576 (37%), Gaps = 148/576 (25%)
Query: 90 KKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGD 149
K+ + ++AINFV +KNRF Y +FLDIL Y+ + +PI VY +VS LF + D
Sbjct: 344 KQAADLDQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFAEAPD 403
Query: 150 LLEEFTHFLPDTSGAAAAHFASARNPLLRDRS--------SAMTTGRQMHVDKREKTTTL 201
LLEEF FLPD S A A + ++ + SA T + K TT
Sbjct: 404 LLEEFKRFLPDVSVNAPAETQDKSTVVPQESATATPKRSPSATPTSALPPIGKFAPPTTA 463
Query: 202 HADRDLSVDHPDPELDRG--VMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGNLE 259
A P PE RG ++T +R EE R N LE
Sbjct: 464 KA-------QPAPEKRRGEPAVQTRNHSKRTRTATSSVEETTPRAFNVPIAQNKNPSELE 516
Query: 260 RLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEK 319
L H + +
Sbjct: 517 FLEHAR----------------------------------------------------QY 524
Query: 320 LKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLA 379
L N Y EF+K L +YS+E+ + L+ G +M D V N
Sbjct: 525 LANESKYNEFIKLLELYSQEVFDKNALVERCYVFFGSNEHLMNWLKDLVKYNPAN----- 579
Query: 380 GVMNKKSLWNEGHGQKPLKVEEKDRDRGRDDGVKERDRELRERDKSTGISNKDVSIPKVS 439
P+ V R R + KS G S + +PK+
Sbjct: 580 ----------------PIPV--------------PRPRVDLTQCKSCGPSYR--LLPKIE 607
Query: 440 LSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSF 499
L + + + WT L A VS+P T SED F
Sbjct: 608 LLLPCSGRDD---LCWTILN-----DAWVSFP-------------------TLASEDSGF 640
Query: 500 KHMRKNQYEESLFRCEDDRFELD---------MLLESINATSKKVEEIIEKVNDDIIPGD 550
RKNQ+EE+L + E++R+E D + L I+A +EK N
Sbjct: 641 IAHRKNQFEENLHKLEEERYEYDRHIGANMRFIELLQIHADKMLKMSEVEKAN-----WT 695
Query: 551 IPIRIEEHLSALNLRCIERLYG-DHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCRE 609
+P + ++ + I+++YG +H ++E L+KN S+ +P++L RLK+K EW +
Sbjct: 696 LPSNLGGKSVSIYHKVIKKVYGKEHAQQIIENLQKNPSVTIPIVLERLKKKDREWRSLQN 755
Query: 610 DFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
+N++W +I KN+++SLDH+ FK D KS + K
Sbjct: 756 HWNELWHDIEEKNFYRSLDHQGVSFKSVDKKSTTPK 791
Score = 87.0 bits (214), Expect = 1e-16
Identities = 42/80 (52%), Positives = 57/80 (70%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
++L DALSYL V+ F + E Y+EFL++MK+FK+Q I+T VI RV +LF G+ +L
Sbjct: 178 RQLNVTDALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAGYPNL 237
Query: 65 ILGFNTFLPKGYAITLPSDD 84
I GFNTFLP GY+I + S D
Sbjct: 238 IQGFNTFLPPGYSIEISSAD 257
Score = 45.1 bits (105), Expect = 6e-04
Identities = 24/101 (23%), Positives = 46/101 (44%), Gaps = 10/101 (9%)
Query: 87 PLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQD 146
P + + + +A++++ +K +F +Y FLDI+ ++ + V VS LF
Sbjct: 174 PQEYRQLNVTDALSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAG 233
Query: 147 HGDLLEEFTHFLP----------DTSGAAAAHFASARNPLL 177
+ +L++ F FLP D A H + + PL+
Sbjct: 234 YPNLIQGFNTFLPPGYSIEISSADPGSLAGIHITTPQGPLM 274
Score = 39.3 bits (90), Expect = 0.034
Identities = 22/90 (24%), Positives = 44/90 (48%), Gaps = 1/90 (1%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
Q + A++++ V+ F + E Y+ FL+++K ++ + V +V +LF DL
Sbjct: 345 QAADLDQAINFVNNVKNRFSHKPEAYNSFLDILKSYQHDQRPIQLVYFQVSQLFAEAPDL 404
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVE 94
+ F FLP ++ P++ + P E
Sbjct: 405 LEEFKRFLP-DVSVNAPAETQDKSTVVPQE 433
Score = 34.7 bits (78), Expect = 0.83
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query: 310 LAFLEKVKEKL-KNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
L++L+ VK + + P+ Y EFL + + + I E++ V L Y ++++GF+ F+
Sbjct: 186 LSYLDLVKLQFHQEPEIYNEFLDIMKEFKSQAIETPEVITRVSKLFAGYPNLIQGFNTFL 245
>SIN3_YEAST (P22579) Paired amphipathic helix protein SIN3
Length = 1536
Score = 117 bits (292), Expect = 1e-25
Identities = 98/342 (28%), Positives = 156/342 (44%), Gaps = 67/342 (19%)
Query: 310 LAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVT 369
+ F EK K + N Y EFLK L++YS+ DI++ DD V
Sbjct: 666 VTFFEKAKRYIGNKHLYTEFLKILNLYSQ--------------------DILD-LDDLVE 704
Query: 370 QCEKNEGFLAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRDDGVKERDRELRERDKSTGIS 429
+ + F G + W + V +++ + ++ V E+ R + ++ G S
Sbjct: 705 KVD----FYLGSNKELFTWFKNF------VGYQEKTKCIENIVHEKHRLDLDLCEAFGPS 754
Query: 430 NKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPKIQKTELGAKVLNDHWVS 489
K L K T M P + ++ +VLND WV
Sbjct: 755 YK-------RLPKSDTFM-----------------------PCSGRDDMCWEVLNDEWVG 784
Query: 490 -VTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIP 548
SED F RKNQYEE+LF+ E++R E D +ES T + +E I+ K+ +
Sbjct: 785 HPVWASEDSGFIAHRKNQYEETLFKIEEERHEYDFYIESNLRTIQCLETIVNKIENMTEN 844
Query: 549 GDIPIRIEEHLSALNL----RCIERLYG-DHGLDVMEVLKKNASLALPVILTRLKQKQEE 603
++ L ++ + I ++Y + G ++++ L ++ ++ PV+L RLKQK EE
Sbjct: 845 EKANFKLPPGLGHTSMTIYKKVIRKVYDKERGFEIIDALHEHPAVTAPVVLKRLKQKDEE 904
Query: 604 WARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
W R + ++NKVW E+ K + KSLDH FKQ D K L+TK
Sbjct: 905 WRRAQREWNKVWRELEQKVFFKSLDHLGLTFKQADKKLLTTK 946
Score = 90.9 bits (224), Expect = 1e-17
Identities = 44/83 (53%), Positives = 59/83 (71%)
Query: 7 LTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLIL 66
L DALSYL+ V+ F + + Y+ FL++MKDFK+Q IDT GVIERV LF+G+ LI
Sbjct: 219 LNVKDALSYLEQVKFQFSSRPDIYNLFLDIMKDFKSQAIDTPGVIERVSTLFRGYPILIQ 278
Query: 67 GFNTFLPKGYAITLPSDDEQPLQ 89
GFNTFLP+GY I S+ + P++
Sbjct: 279 GFNTFLPQGYRIECSSNPDDPIR 301
Score = 81.3 bits (199), Expect = 8e-15
Identities = 41/86 (47%), Positives = 58/86 (66%), Gaps = 2/86 (2%)
Query: 80 LPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQE 139
+P +D + + VEF +AI++V KIK RF +YK FL+IL Y++E KPI VY +
Sbjct: 395 VPQEDAK--KNVDVEFSQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQKPINEVYAQ 452
Query: 140 VSALFQDHGDLLEEFTHFLPDTSGAA 165
V+ LFQ+ DLLE+F FLPD+S +A
Sbjct: 453 VTHLFQNAPDLLEDFKKFLPDSSASA 478
Score = 54.3 bits (129), Expect = 1e-06
Identities = 60/281 (21%), Positives = 113/281 (39%), Gaps = 29/281 (10%)
Query: 91 KPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDL 150
+P+ ++A++++ ++K +F +Y FLDI+ ++ + V + VS LF+ + L
Sbjct: 217 RPLNVKDALSYLEQVKFQFSSRPDIYNLFLDIMKDFKSQAIDTPGVIERVSTLFRGYPIL 276
Query: 151 LEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMHVDKREKTTTLHADRDLSVD 210
++ F FLP +S + +R + TT ++ + TT D
Sbjct: 277 IQGFNTFLPQ---GYRIECSSNPDDPIRVTTPMGTTTVNNNISPSGRGTT-----DAQEL 328
Query: 211 HPDPELD-RGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGNLERLPHKKKSVH 269
PE D GV + + +D++ + LP S
Sbjct: 329 GSFPESDGNGVQQPSNVPMVPSSVYQSEQNQDQQ---------------QSLPLLATSSG 373
Query: 270 RATDPGTAEPLH-DADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKN-PDDYQ 327
+ P H + L+P + K ++ S ++++ K+K + + PD Y+
Sbjct: 374 LPSIQQPEMPAHRQIPQSQSLVPQE---DAKKNVDVEFSQAISYVNKIKTRFADQPDIYK 430
Query: 328 EFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFV 368
FL+ L Y RE E+ A V L D++E F F+
Sbjct: 431 HFLEILQTYQREQKPINEVYAQVTHLFQNAPDLLEDFKKFL 471
Score = 40.4 bits (93), Expect = 0.015
Identities = 19/68 (27%), Positives = 36/68 (52%)
Query: 10 NDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFN 69
+ A+SY+ ++ F + + Y FLE+++ ++ ++ V +V LF+ DL+ F
Sbjct: 409 SQAISYVNKIKTRFADQPDIYKHFLEILQTYQREQKPINEVYAQVTHLFQNAPDLLEDFK 468
Query: 70 TFLPKGYA 77
FLP A
Sbjct: 469 KFLPDSSA 476
Score = 35.8 bits (81), Expect = 0.37
Identities = 19/73 (26%), Positives = 38/73 (52%), Gaps = 2/73 (2%)
Query: 86 QPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQ 145
+P++ + E + F K K R+ GN +Y FL ILN+Y +++ + + ++V
Sbjct: 654 EPIENN-ISLNEEVTFFEKAK-RYIGNKHLYTEFLKILNLYSQDILDLDDLVEKVDFYLG 711
Query: 146 DHGDLLEEFTHFL 158
+ +L F +F+
Sbjct: 712 SNKELFTWFKNFV 724
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 104 bits (259), Expect = 9e-22
Identities = 58/136 (42%), Positives = 85/136 (61%), Gaps = 12/136 (8%)
Query: 7 LTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLIL 66
+TT DAL+YLKAV++ F+ D EKYD FLEV+ D K Q +DT+GVI R+K+LFKGH DL+L
Sbjct: 304 VTTGDALNYLKAVKDKFE-DSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLL 362
Query: 67 GFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMY 126
GFNT+L K Y IT+ +D+ P I+F+ K++ ++ + +T NM
Sbjct: 363 GFNTYLSKEYQITILPEDDFP-----------IDFLDKVEGPYEMTYQQAQTVQANANMQ 411
Query: 127 RKELKPITAVYQEVSA 142
+ P ++ Q S+
Sbjct: 412 PQTEYPSSSAVQSFSS 427
Score = 39.7 bits (91), Expect = 0.026
Identities = 19/62 (30%), Positives = 38/62 (60%), Gaps = 1/62 (1%)
Query: 97 EAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTH 156
+A+N++ +K++F+ +++ Y TFL++LN + + + V + LF+ H DLL F
Sbjct: 308 DALNYLKAVKDKFEDSEK-YDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 157 FL 158
+L
Sbjct: 367 YL 368
Score = 33.5 bits (75), Expect = 1.9
Identities = 15/61 (24%), Positives = 36/61 (58%)
Query: 310 LAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVT 369
L +L+ VK+K ++ + Y FL+ L+ + + ++A + DL + D++ GF+ +++
Sbjct: 310 LNYLKAVKDKFEDSEKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNTYLS 369
Query: 370 Q 370
+
Sbjct: 370 K 370
>PST2_SCHPO (O13919) Paired amphipathic helix protein pst2 (SIN3
homolog 2)
Length = 1075
Score = 99.4 bits (246), Expect = 3e-20
Identities = 56/170 (32%), Positives = 97/170 (56%), Gaps = 6/170 (3%)
Query: 482 VLNDHWVS-VTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIE 540
+LND WVS T SE+ F RK YEE++ + E++R+E D +E+ + T K +++I
Sbjct: 363 ILNDDWVSHPTWASEESGFIVQRKTPYEEAMTKLEEERYEFDRHIEATSWTIKSLKKIQN 422
Query: 541 KVNDDIIPGDIPIRIEEHLS----ALNLRCIERLY-GDHGLDVMEVLKKNASLALPVILT 595
++N+ +EE L ++ + I+ +Y +H ++ + L++ L LP++++
Sbjct: 423 RINELPEEERETYTLEEGLGLPSKSIYKKTIKLVYTSEHAEEMFKALERMPCLTLPLVIS 482
Query: 596 RLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
RL++K EEW + W I KNY KSLD + YFK +D K++S+K
Sbjct: 483 RLEEKNEEWKSVKRSLQPGWRSIEFKNYDKSLDSQCVYFKARDKKNVSSK 532
Score = 57.8 bits (138), Expect = 9e-08
Identities = 38/178 (21%), Positives = 72/178 (40%), Gaps = 20/178 (11%)
Query: 4 GQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKD 63
G L D ++++ + + + Y F++V+K + +D G IER+ + + + D
Sbjct: 31 GTALPELDIKAFVQKLGQRLCHRPYVYSAFMDVVKALHNEIVDFPGFIERISVILRDYPD 90
Query: 64 LILGFNTFLPKGY--------------------AITLPSDDEQPLQKKPVEFEEAINFVG 103
L+ N FLP Y ++ PS P + AI FV
Sbjct: 91 LLEYLNIFLPSSYKYLLSNSGANFTLQFTTPSGPVSTPSTYVATYNDLPCTYHRAIGFVS 150
Query: 104 KIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT 161
+++ N + D L ++ ++ + V++L +H L EF +FLP +
Sbjct: 151 RVRRALLSNPEQFFKLQDSLRKFKNSECSLSELQTIVTSLLAEHPSLAHEFHNFLPSS 208
>CTF8_HUMAN (Q96D72) Protein C20orf158
Length = 478
Score = 45.1 bits (105), Expect = 6e-04
Identities = 42/124 (33%), Positives = 55/124 (43%), Gaps = 25/124 (20%)
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHP-----------DPELDRGVM--RTD 224
R+R R+ D R + H D+D S D + E DRG R+
Sbjct: 353 RERERGRNWSRERDWD-RPREWDRHRDKDSSRDWDRNRERSANRDREREADRGKEWDRSR 411
Query: 225 KEQRRREREKDRREERDR---RERERD---DRD-----YDNDGNLERLPHKKKSVHRATD 273
+ R RERE+DRR +RDR RER+RD DR+ D + E K RA+D
Sbjct: 412 ERSRNRERERDRRRDRDRSRSRERDRDKARDRERGRDRKDRSKSKESARDPKPEASRASD 471
Query: 274 PGTA 277
GTA
Sbjct: 472 AGTA 475
>RED_HUMAN (Q13123) Red protein (RER protein) (IK factor) (Cytokine
IK)
Length = 557
Score = 43.5 bits (101), Expect = 0.002
Identities = 71/266 (26%), Positives = 110/266 (40%), Gaps = 36/266 (13%)
Query: 194 KREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRR-REREKDRREERDR-RERERD-DR 250
K E+ AD ++ D D DKE+ R RERE+DR +RDR RERER+ DR
Sbjct: 305 KLEEKKPPEADMNIFEDIGDYVPSTTKTPRDKERERYRERERDRERDRDRDRERERERDR 364
Query: 251 DYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVL 310
+ + + + ER KK+ + EP+ +D P S+ K +KS+
Sbjct: 365 ERERERDREREEEKKRHSYFEKPKVDDEPM-----DVDKGPGST----KELIKSINEKFA 415
Query: 311 AFLE-KVKEKLKNPDDYQ---EFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFD- 365
+ E LK P+D + +F + Y+ + +A+ D Y+ + +G
Sbjct: 416 GSAGWEGTESLKKPEDKKQLGDFFGMSNSYAECYPATMDDMAVDSDEEVDYSKMDQGNKK 475
Query: 366 ------DFVTQCEKNEGFLAGVMNKKSLWNEGHGQKPLKVEE--KDRDRGRDDGVKERDR 417
DF TQ E +E MN K + Q +K+ E K R + E DR
Sbjct: 476 GPLGRWDFDTQEEYSE-----YMNNKEALPKAAFQYGIKMSEGRKTRRFKETNDKAELDR 530
Query: 418 ELR------ERDKSTGISNKDVSIPK 437
+ + E+ K +V PK
Sbjct: 531 QWKKISAIIEKRKKMEADGVEVKRPK 556
>RU17_XENLA (P09406) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70) (U1-70K)
Length = 471
Score = 43.1 bits (100), Expect = 0.002
Identities = 31/95 (32%), Positives = 51/95 (53%), Gaps = 7/95 (7%)
Query: 187 GRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRR----REREKDRREERD- 241
G ++ + EK+ +RD + + + D+ R D+++RR R+REKDR +RD
Sbjct: 338 GIELKQEPEEKSRERDRERDRDREKGEKDRDKDRDR-DRDRRRSHRDRDREKDRDRDRDR 396
Query: 242 RRERERD-DRDYDNDGNLERLPHKKKSVHRATDPG 275
RR+R+RD +RD D+ +R +K R D G
Sbjct: 397 RRDRDRDRERDKDHKRERDRGDRSEKREERVPDNG 431
Score = 40.8 bits (94), Expect = 0.012
Identities = 26/75 (34%), Positives = 37/75 (48%), Gaps = 6/75 (8%)
Query: 213 DPELDRGVMRTDKEQRRREREKDRREERDRRERERD-DRDYDNDGNLERLPHKKKSVHRA 271
D E DR D+E+ ++R+KDR +RDRR RD DR+ D D + +R + + R
Sbjct: 353 DRERDR-----DREKGEKDRDKDRDRDRDRRRSHRDRDREKDRDRDRDRRRDRDRDRERD 407
Query: 272 TDPGTAEPLHDADEK 286
D D EK
Sbjct: 408 KDHKRERDRGDRSEK 422
Score = 40.4 bits (93), Expect = 0.015
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 179 DRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRRE 238
++S R +K EK DRD + DR R RRR+R++DR
Sbjct: 347 EKSRERDRERDRDREKGEKDRDKDRDRDRDRRRSHRDRDREKDRDRDRDRRRDRDRDRER 406
Query: 239 ERD-RRERERDDRDYDNDGNLERLPHKKKSVHRATD 273
++D +RER+R DR + ER+P + +A +
Sbjct: 407 DKDHKRERDRGDRSEKRE---ERVPDNGMVMEQAEE 439
Score = 39.3 bits (90), Expect = 0.034
Identities = 51/212 (24%), Positives = 91/212 (42%), Gaps = 33/212 (15%)
Query: 217 DRGVMRTDKEQRRREREKDRREERDR----RERERDDRDYDNDGNLERLPHKKKSVHRAT 272
D + + ++ R E+DR ERDR RERE++ R+ + ER K +S +
Sbjct: 208 DVNIRHSGRDDTSRYDERDRERERDRRERSREREKEPRERRRSRSRER-RRKSRSREKEE 266
Query: 273 DPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKC 332
T E D D++ D ++ + K +S E+ +E+ ++ + +E ++
Sbjct: 267 RKRTREKSKDKDKEKD--KDNKDRDRKRRSRS--------RERKRERDRDREKKEERVEA 316
Query: 333 LHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNKKSLWNEGH 392
+ + + + A +GDL G D + E E ++ +
Sbjct: 317 -EVPEADDAPQDD--AQIGDL---------GIDGIELKQEPEE-----KSRERDRERDRD 359
Query: 393 GQKPLKVEEKDRDRGRDDGVKERDRELRERDK 424
+K K +KDRDR RD RDR+ RE+D+
Sbjct: 360 REKGEKDRDKDRDRDRDRRRSHRDRD-REKDR 390
Score = 37.0 bits (84), Expect = 0.17
Identities = 28/96 (29%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDR- 236
R+R R+ R+K DR S D E DR R + R R+RE+D+
Sbjct: 350 RERDRERDRDREKGEKDRDKDRDRDRDRRRSHRDRDREKDRDRDRDRRRDRDRDRERDKD 409
Query: 237 -REERDR--RERERDDRDYDNDGNLERLPHKKKSVH 269
+ ERDR R +R++R DN +E+ + ++
Sbjct: 410 HKRERDRGDRSEKREERVPDNGMVMEQAEETSQDMY 445
Score = 36.6 bits (83), Expect = 0.22
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Query: 180 RSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREE 239
RS + R+ D+ +K + A+ + D P + G + D + ++E E+ RE
Sbjct: 293 RSRSRERKRERDRDREKKEERVEAEVPEADDAPQDDAQIGDLGIDGIELKQEPEEKSRER 352
Query: 240 RDRRERERDDRDYDNDGNLERLPHKKKSVHRATD 273
R+R+R+ + D D + +R +++S HR D
Sbjct: 353 DRERDRDREKGEKDRDKDRDRDRDRRRS-HRDRD 385
Score = 34.7 bits (78), Expect = 0.83
Identities = 22/79 (27%), Positives = 40/79 (49%), Gaps = 8/79 (10%)
Query: 173 RNPLLRDRSSAMTTGRQMHV-DKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRRE 231
+ P R RS + R+ +K E+ T +D D E D+ D++++RR
Sbjct: 242 KEPRERRRSRSRERRRKSRSREKEERKRTREKSKD-----KDKEKDKD--NKDRDRKRRS 294
Query: 232 REKDRREERDRRERERDDR 250
R ++R+ ERDR ++++R
Sbjct: 295 RSRERKRERDRDREKKEER 313
Score = 33.1 bits (74), Expect = 2.4
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 4/87 (4%)
Query: 188 RQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREK----DRREERDRR 243
R+ D+RE++ + E R +KE+R+R REK D+ +++D +
Sbjct: 227 RERERDRRERSREREKEPRERRRSRSRERRRKSRSREKEERKRTREKSKDKDKEKDKDNK 286
Query: 244 ERERDDRDYDNDGNLERLPHKKKSVHR 270
+R+R R + ER ++K R
Sbjct: 287 DRDRKRRSRSRERKRERDRDREKKEER 313
>RED_MOUSE (Q9Z1M8) Red protein (RER protein)
Length = 557
Score = 43.1 bits (100), Expect = 0.002
Identities = 62/227 (27%), Positives = 96/227 (41%), Gaps = 28/227 (12%)
Query: 194 KREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRR-REREKDRREERDR-RERERD-DR 250
K E+ AD ++ D D DKE+ R RERE+DR +RDR R+RERD +R
Sbjct: 305 KLEEKKPPEADMNIFEDIGDYVPSTTKTPRDKERERYRERERDRERDRDRERDRERDRER 364
Query: 251 DYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCSPVL 310
+ + D ER KK+ + EP+ +D P S+ K +KS+
Sbjct: 365 ERERDREREREEEKKRHSYFEKPKVDDEPM-----DVDKGPGSA----KELIKSINEKFA 415
Query: 311 AFLE-KVKEKLKNPDDYQ---EFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFD- 365
+ E LK P+D + +F + Y+ + +A+ D Y+ + +G
Sbjct: 416 GSAGWEGTESLKKPEDKKQLGDFFGMSNSYAECYPATMDDMAVDSDEEVDYSKMDQGNKK 475
Query: 366 ------DFVTQCEKNEGFLAGVMNKKSLWNEGHGQKPLKVEEKDRDR 406
DF TQ E +E MN K + Q +K+ E + R
Sbjct: 476 GPLGRWDFDTQEEYSE-----YMNNKEALPKAAFQYGIKMSEGRKTR 517
>NLFE_HUMAN (P18615) Negative elongation factor E (NELF-E) (RD
protein)
Length = 380
Score = 42.0 bits (97), Expect = 0.005
Identities = 29/83 (34%), Positives = 44/83 (52%), Gaps = 7/83 (8%)
Query: 179 DRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRRE 238
+RS A ++ R+++ + DRD D E DR R +R R+R++DR
Sbjct: 172 ERSGAHSSASPPRSRSRDRSHERNRDRDR-----DRERDRDRDRDRDRERDRDRDRDRDR 226
Query: 239 ERDR-RERERDDRDYDNDGNLER 260
+R+R R+RER DRD D +G R
Sbjct: 227 DRERDRDRER-DRDRDREGPFRR 248
Score = 36.2 bits (82), Expect = 0.29
Identities = 22/65 (33%), Positives = 33/65 (49%), Gaps = 2/65 (3%)
Query: 222 RTDKEQRRREREKDRREERDR-RERERD-DRDYDNDGNLERLPHKKKSVHRATDPGTAEP 279
R+ R R++DR ERDR R+R+RD +RD D D + +R + + R D P
Sbjct: 186 RSRDRSHERNRDRDRDRERDRDRDRDRDRERDRDRDRDRDRDRERDRDRERDRDRDREGP 245
Query: 280 LHDAD 284
+D
Sbjct: 246 FRRSD 250
Score = 32.0 bits (71), Expect = 5.4
Identities = 20/48 (41%), Positives = 26/48 (53%), Gaps = 4/48 (8%)
Query: 230 REREKDRREERDR---RERERD-DRDYDNDGNLERLPHKKKSVHRATD 273
R R +DR ER+R R+RERD DRD D D +R + + R D
Sbjct: 184 RSRSRDRSHERNRDRDRDRERDRDRDRDRDRERDRDRDRDRDRDRERD 231
>ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1338
Score = 42.0 bits (97), Expect = 0.005
Identities = 23/58 (39%), Positives = 34/58 (57%), Gaps = 1/58 (1%)
Query: 215 ELDRGVMRTDKEQRRREREKDRREERDRRERERD-DRDYDNDGNLERLPHKKKSVHRA 271
E +R + R + + RERE+DR ERDR +RER+ +RD D +R K+ S R+
Sbjct: 1269 ERNRQLEREKRREHSRERERDRERERDRGDRERERERDRDRGRERDRRDTKRHSRSRS 1326
Score = 35.8 bits (81), Expect = 0.37
Identities = 16/70 (22%), Positives = 35/70 (49%)
Query: 215 ELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGNLERLPHKKKSVHRATDP 274
E ++ ++E+RR+ERE++ ++ER++ +R + + E +++ R D
Sbjct: 1237 EAEQAERAKEREKRRKEREEEEQKEREKEAERERNRQLEREKRREHSRERERDRERERDR 1296
Query: 275 GTAEPLHDAD 284
G E + D
Sbjct: 1297 GDRERERERD 1306
Score = 35.4 bits (80), Expect = 0.49
Identities = 46/212 (21%), Positives = 83/212 (38%), Gaps = 33/212 (15%)
Query: 225 KEQRRREREK--DRREERDRRERERDDRDYDNDGNLE--RLPHKKKSVHRATDPGTAEPL 280
+EQ R RE+ +R E +RRER R +R++D D E R + + R + E
Sbjct: 1135 REQERAVREQWAEREREMERRERTRSEREWDRDKVREGPRSRSRSRDRRRKERAKSKEKK 1194
Query: 281 HDADEKLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREI 340
+ EK P + +D K+ +P + +L + ++ +E + RE
Sbjct: 1195 SEKKEKAQEEPPAKLLDDLFR-KTKAAPCIYWLPLTESQIVQ----KEAEQAERAKEREK 1249
Query: 341 ITRQELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNKKSLWNEGHGQKPLKVE 400
++ + E+ E + ++ +
Sbjct: 1250 RRKER-----------------------EEEEQKEREKEAERERNRQLEREKRREHSRER 1286
Query: 401 EKDRDRGRDDGVKERDRELRERDKSTGISNKD 432
E+DR+R RD G +ER+RE R+RD+ +D
Sbjct: 1287 ERDRERERDRGDRERERE-RDRDRGRERDRRD 1317
Score = 32.7 bits (73), Expect = 3.2
Identities = 25/63 (39%), Positives = 34/63 (53%), Gaps = 18/63 (28%)
Query: 188 RQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERER 247
R+ H +RE+ DR + E DRG D+E R RER++DR ERDRR+ +R
Sbjct: 1279 RREHSRERER------DR-------ERERDRG----DRE-RERERDRDRGRERDRRDTKR 1320
Query: 248 DDR 250
R
Sbjct: 1321 HSR 1323
Score = 32.3 bits (72), Expect = 4.1
Identities = 27/101 (26%), Positives = 41/101 (39%), Gaps = 9/101 (8%)
Query: 189 QMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDR-RERER 247
Q ++ E+ R + E ++ R Q RE+ ++ ER+R RERER
Sbjct: 1235 QKEAEQAERAKEREKRRKEREEEEQKEREKEAERERNRQLEREKRREHSRERERDRERER 1294
Query: 248 D------DRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHD 282
D +R+ D D ER K R+ T P+ D
Sbjct: 1295 DRGDRERERERDRDRGRERDRRDTKRHSRSRSRST--PVRD 1333
Score = 32.0 bits (71), Expect = 5.4
Identities = 24/81 (29%), Positives = 39/81 (47%), Gaps = 13/81 (16%)
Query: 179 DRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDR-- 236
+R+ R+ ++ +K A+R+ + +L+R R +R R+RE++R
Sbjct: 1242 ERAKEREKRRKEREEEEQKEREKEAERERN-----RQLEREKRREHSRERERDRERERDR 1296
Query: 237 -----REERDR-RERERDDRD 251
ERDR R RERD RD
Sbjct: 1297 GDRERERERDRDRGRERDRRD 1317
>YD35_MYCPN (P75443) Hypothetical protein MPN335 (F10_orf741)
Length = 741
Score = 40.8 bits (94), Expect = 0.012
Identities = 45/215 (20%), Positives = 91/215 (41%), Gaps = 24/215 (11%)
Query: 220 VMRTDKEQRRREREKDRREERDRRERER-DDRDYDNDGNLERLPHKKKSVHRATDPGTAE 278
++++ ++ E+E+ +EE+++ E++ + +D + ++ P+ K S
Sbjct: 396 IIQSFRDYEAEEKERQEKEEKEKAEKDNGNGQDSNKVNSVSTEPNNKNS----------- 444
Query: 279 PLHDADEKLDLLPNSSTCEDKSSLKSLCSPVL-AFLEKVK-EKLKNPDDYQEFLKCLHIY 336
DAD K + + S +D S K P L F ++V K + LK +H
Sbjct: 445 --SDADSKDNNDSSDSQGKDSSKSKPKFRPRLPQFFDRVSINYSKFSSSFNTQLKHIH-- 500
Query: 337 SREIITRQELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNKKSLWNEGHGQKP 396
+ + TR E + + D + + + +D+F Q K+ G + K QK
Sbjct: 501 -QGLSTRDEAMEIFKDKVALFYALSMAYDNFTFQ--KDSGIIDNTSLNKFKTEFNKKQKE 557
Query: 397 LKVEEKDRDRGRDDGVKERDRELRERDKSTGISNK 431
L + K + G+D ++ R + DK+ + K
Sbjct: 558 LTEKNKQKQDGKD---QKSQRMTQGADKAVTVEPK 589
>SFRC_RAT (Q9JKL7) Splicing factor, arginine/serine-rich 12
(Serine-arginine-rich splicing regulatory protein 86)
(SRrp86) (SR-related protein of 86 kDa)
Length = 494
Score = 40.8 bits (94), Expect = 0.012
Identities = 28/100 (28%), Positives = 50/100 (50%), Gaps = 3/100 (3%)
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHP-DPELDRGVMRTDKEQRRREREKDR 236
R +S + + R D REK +R+ + + E ++ R + + RE+EKD
Sbjct: 251 RRKSRSRSRSRDKRKDTREKVKERVKEREREKEKEREKEREKDKERGKNKDKDREKEKDH 310
Query: 237 REERDR-RERERD-DRDYDNDGNLERLPHKKKSVHRATDP 274
+ERD+ +E+E+D D++ + D + E +KK T P
Sbjct: 311 EKERDKEKEKEQDKDKEREKDRSKEADEKRKKEKKSRTPP 350
Score = 36.2 bits (82), Expect = 0.29
Identities = 23/93 (24%), Positives = 48/93 (50%), Gaps = 3/93 (3%)
Query: 194 KREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYD 253
+R K+ + RD D + +R ++ + ++ +EREK+R ++++R + + DR+ +
Sbjct: 250 ERRKSRSRSRSRDKRKDTREKVKER--VKEREREKEKEREKEREKDKERGKNKDKDREKE 307
Query: 254 NDGNLERLPHKKKSVHRATDPGTAEPLHDADEK 286
D ER K+K + + + +ADEK
Sbjct: 308 KDHEKERDKEKEKEQDKDKE-REKDRSKEADEK 339
Score = 33.1 bits (74), Expect = 2.4
Identities = 34/121 (28%), Positives = 47/121 (38%), Gaps = 19/121 (15%)
Query: 174 NPLLRDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRRERE 233
N R RS++ R+ T + S P P RG + ++ ++RE
Sbjct: 354 NSSRRSRSASRERRRRRSRSSSRSPRTSKTIKRKSSRSPSP---RG-----RNKKEKKRE 405
Query: 234 KDRREERDRRERER--------DDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADE 285
K+R DRRERER DRD P K+K + D + DADE
Sbjct: 406 KERDHISDRRERERSTSTKKSSSDRDGKEKVEKTNTPVKEKEHSKEADSTVST---DADE 462
Query: 286 K 286
K
Sbjct: 463 K 463
>DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-dependent
RNA helicase
Length = 1168
Score = 40.8 bits (94), Expect = 0.012
Identities = 29/106 (27%), Positives = 46/106 (43%), Gaps = 7/106 (6%)
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRR 237
+D+ + + ++R + + DRD D + DR R D+ R R+RE++R
Sbjct: 104 KDKVKELEKEIEREAEERRREEDRNRDRDRRESGRDRDRDRNRDRDDRRDRHRDRERNRG 163
Query: 238 EER------DRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTA 277
+E DRR RER D +G R + K + D G A
Sbjct: 164 DEEGEDRRSDRRHRERGRGD-GGEGEDRRRDRRAKDEYVEEDKGGA 208
Score = 33.9 bits (76), Expect = 1.4
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query: 193 DKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERD-RRERERD-DR 250
D ++K L + + + E DR R D+ + R+R++DR +RD RR+R RD +R
Sbjct: 102 DTKDKVKELEKEIEREAEERRREEDRNRDR-DRRESGRDRDRDRNRDRDDRRDRHRDRER 160
Query: 251 DYDNDGNLERLPHKKKSVHRATDPGTAE 278
+ ++ +R ++ D G E
Sbjct: 161 NRGDEEGEDRRSDRRHRERGRGDGGEGE 188
>TRHY_RABIT (P37709) Trichohyalin
Length = 1407
Score = 40.4 bits (93), Expect = 0.015
Identities = 23/85 (27%), Positives = 46/85 (54%), Gaps = 3/85 (3%)
Query: 194 KREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDYD 253
++E+ A+ + + L+R + + ++++RRRERE+ REE+ RR++E + R
Sbjct: 1302 RQERDRRYRAEEQFAREEKSRRLERELRQEEEQRRRRERERKFREEQLRRQQEEEQR--- 1358
Query: 254 NDGNLERLPHKKKSVHRATDPGTAE 278
ER + +S + +PGT +
Sbjct: 1359 RRQLRERQFREDQSRRQVLEPGTRQ 1383
Score = 35.4 bits (80), Expect = 0.49
Identities = 16/40 (40%), Positives = 25/40 (62%)
Query: 213 DPELDRGVMRTDKEQRRREREKDRREERDRRERERDDRDY 252
+ EL R R ++E+ RE EK +REE ++R R+ +R Y
Sbjct: 550 EEELQREKRRQEREREYREEEKLQREEDEKRRRQERERQY 589
Score = 34.3 bits (77), Expect = 1.1
Identities = 15/34 (44%), Positives = 23/34 (67%)
Query: 215 ELDRGVMRTDKEQRRREREKDRREERDRRERERD 248
E DR ++++RR+ERE+ R ERDR+ RE +
Sbjct: 1071 ERDRKFREEEQQRRRQEREQQLRRERDRKFREEE 1104
Score = 31.2 bits (69), Expect = 9.2
Identities = 20/78 (25%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Query: 173 RNPLLRDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPEL--DRGVMRTDKEQRRR 230
R LR+ + + + ++E+ L + L + + EL +R ++EQ R
Sbjct: 1166 RERKLREEEQLLQEREEERLRRQERARKLREEEQL-LRQEEQELRQERARKLREEEQLLR 1224
Query: 231 EREKDRREERDRRERERD 248
+ E++ R+ERDR+ RE +
Sbjct: 1225 QEEQELRQERDRKFREEE 1242
Score = 31.2 bits (69), Expect = 9.2
Identities = 23/77 (29%), Positives = 39/77 (49%), Gaps = 6/77 (7%)
Query: 176 LLRDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPEL--DRGVMRTDKEQR--RRE 231
LLR + R+ + RE+ L + L D +L + +++ +E+R R+E
Sbjct: 704 LLRQEEQELRQERERKL--REEEQLLRREEQLLRQERDRKLREEEQLLQESEEERLRRQE 761
Query: 232 REKDRREERDRRERERD 248
RE+ R ERDR+ RE +
Sbjct: 762 REQQLRRERDRKFREEE 778
>YP68_CAEEL (Q09217) Hypothetical protein B0495.8 in chromosome II
Length = 313
Score = 40.0 bits (92), Expect = 0.020
Identities = 18/44 (40%), Positives = 25/44 (55%)
Query: 217 DRGVMRTDKEQRRREREKDRREERDRRERERDDRDYDNDGNLER 260
DRG R D+++ RR R++ R +R R +R DR YD D R
Sbjct: 264 DRGEYRGDRDRDRRNRDRSRSRDRSYRRDDRGDRRYDRDNRDRR 307
>THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2)
Length = 1478
Score = 40.0 bits (92), Expect = 0.020
Identities = 28/110 (25%), Positives = 55/110 (49%), Gaps = 12/110 (10%)
Query: 207 LSVDHPDPELDRGVMRT-DKEQ----RRREREKDRREERDRRERERDDRDYDNDGNLERL 261
L ++H P L + R DK+ R R RE+++++E+DR+ER+RD + D + +
Sbjct: 1323 LYINHTPPPLSKSKEREMDKKDLDKSRERSREREKKDEKDRKERKRDHSNNDREVPPDLT 1382
Query: 262 PHKKK-------SVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKS 304
+K+ S H++ P + ++ D++ + +S + S KS
Sbjct: 1383 KRRKEENGTMGVSKHKSESPCESPYPNEKDKEKNKSKSSGKEKGSDSFKS 1432
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 80,883,265
Number of Sequences: 164201
Number of extensions: 3684979
Number of successful extensions: 19232
Number of sequences better than 10.0: 268
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 167
Number of HSP's that attempted gapping in prelim test: 15859
Number of HSP's gapped (non-prelim): 1705
length of query: 670
length of database: 59,974,054
effective HSP length: 117
effective length of query: 553
effective length of database: 40,762,537
effective search space: 22541682961
effective search space used: 22541682961
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146588.6


