

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
(395 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
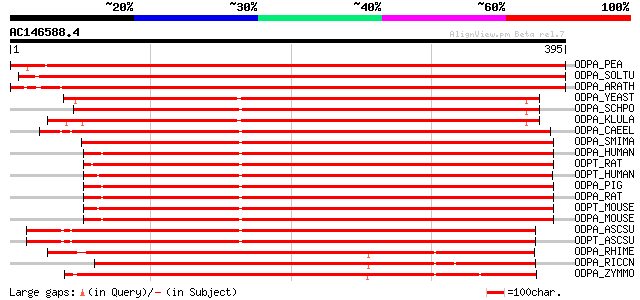
Score E
Sequences producing significant alignments: (bits) Value
ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component alpha subu... 662 0.0
ODPA_SOLTU (P52903) Pyruvate dehydrogenase E1 component alpha su... 649 0.0
ODPA_ARATH (P52901) Pyruvate dehydrogenase E1 component alpha su... 624 e-178
ODPA_YEAST (P16387) Pyruvate dehydrogenase E1 component alpha su... 371 e-102
ODPA_SCHPO (Q10489) Pyruvate dehydrogenase E1 component alpha su... 369 e-102
ODPA_KLULA (O13366) Pyruvate dehydrogenase E1 component alpha su... 365 e-100
ODPA_CAEEL (P52899) Probable pyruvate dehydrogenase E1 component... 365 e-100
ODPA_SMIMA (P52900) Pyruvate dehydrogenase E1 component alpha su... 358 1e-98
ODPA_HUMAN (P08559) Pyruvate dehydrogenase E1 component alpha su... 352 9e-97
ODPT_RAT (Q06437) Pyruvate dehydrogenase E1 component alpha subu... 351 2e-96
ODPT_HUMAN (P29803) Pyruvate dehydrogenase E1 component alpha su... 351 2e-96
ODPA_PIG (P29804) Pyruvate dehydrogenase E1 component alpha subu... 350 3e-96
ODPA_RAT (P26284) Pyruvate dehydrogenase E1 component alpha subu... 350 5e-96
ODPT_MOUSE (P35487) Pyruvate dehydrogenase E1 component alpha su... 349 8e-96
ODPA_MOUSE (P35486) Pyruvate dehydrogenase E1 component alpha su... 349 8e-96
ODPA_ASCSU (P26267) Pyruvate dehydrogenase E1 component alpha su... 345 9e-95
ODPT_ASCSU (P26268) Pyruvate dehydrogenase E1 component alpha su... 340 4e-93
ODPA_RHIME (Q9R9N5) Pyruvate dehydrogenase E1 component, alpha s... 298 1e-80
ODPA_RICCN (Q92IS3) Pyruvate dehydrogenase E1 component, alpha s... 286 8e-77
ODPA_ZYMMO (O66112) Pyruvate dehydrogenase E1 component, alpha s... 279 8e-75
>ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 397
Score = 662 bits (1708), Expect = 0.0
Identities = 322/398 (80%), Positives = 354/398 (88%), Gaps = 4/398 (1%)
Query: 1 MALSRFSSSQF---GSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTT 57
MALSR SSS GS L P+ + + +R ISS +T TLT+ETS+PFT+HNC+PPS +
Sbjct: 1 MALSRLSSSSSSSNGSNLFNPFSAAFTL-NRPISSDTTATLTIETSLPFTAHNCDPPSRS 59
Query: 58 VQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDC 117
V T+ SEL+SFF M LMRRMEIAADSLYKA LIRGFCHLYDGQEAVAVGMEA +KDC
Sbjct: 60 VTTSPSELLSFFRTMALMRRMEIAADSLYKANLIRGFCHLYDGQEAVAVGMEAGTTKKDC 119
Query: 118 VITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQI 177
+ITAYRDHCTFL RGGTL+ V++ELMGR+DGCSKGKGGSMHFY+K+ GFYGGHGIVGAQ+
Sbjct: 120 IITAYRDHCTFLGRGGTLLRVYAELMGRRDGCSKGKGGSMHFYKKDSGFYGGHGIVGAQV 179
Query: 178 PLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGT 237
PLG GLAFGQKY KD +VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGT
Sbjct: 180 PLGCGLAFGQKYLKDESVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGT 239
Query: 238 AEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGH 297
A WRSAKSPAY+KRGDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGH
Sbjct: 240 ATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGH 299
Query: 298 SMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAK 357
SMSDPGSTYRTRDEISGVRQERDPIERVRKL+L+HDI+TEKELKD EKE RK+VDEAIAK
Sbjct: 300 SMSDPGSTYRTRDEISGVRQERDPIERVRKLLLSHDIATEKELKDTEKEVRKEVDEAIAK 359
Query: 358 AKESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
AK+S MPDPSDLF+NVYVKG GVEA G DRKEV+ TLP
Sbjct: 360 AKDSPMPDPSDLFSNVYVKGYGVEAFGVDRKEVRVTLP 397
>ODPA_SOLTU (P52903) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 391
Score = 649 bits (1675), Expect = 0.0
Identities = 314/390 (80%), Positives = 352/390 (89%), Gaps = 3/390 (0%)
Query: 7 SSSQFGSTLLKPYFLSSAI-RHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASEL 65
S+S+ + ++KP LS+A+ R +SS ST T+TVETS+PFTSHN +PPS +V+T+ EL
Sbjct: 4 STSRAINHIMKP--LSAAVCATRRLSSDSTATITVETSLPFTSHNIDPPSRSVETSPKEL 61
Query: 66 MSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDH 125
M+FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI +KDC+ITAYRDH
Sbjct: 62 MTFFKDMTEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDCIITAYRDH 121
Query: 126 CTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAF 185
C FL RGGTLVE F+ELMGR+DGCS+GKGGSMHFY+KE GFYGGHGIVGAQ+PLG+GLAF
Sbjct: 122 CIFLGRGGTLVEAFAELMGRRDGCSRGKGGSMHFYKKESGFYGGHGIVGAQVPLGIGLAF 181
Query: 186 GQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKS 245
QKY K+ VTF +YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKS
Sbjct: 182 AQKYKKEDYVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKS 241
Query: 246 PAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGST 305
PAYYKRGDY PGL+VDGMDV AVKQAC FAK+HALKNGP+ILEMDTYRYHGHSMSDPGST
Sbjct: 242 PAYYKRGDYVPGLRVDGMDVFAVKQACTFAKQHALKNGPIILEMDTYRYHGHSMSDPGST 301
Query: 306 YRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPD 365
YRTRDEISGVRQERDP+ER+R L+LAH+I+TE ELKDIEKE RK VDEAIAKAKES MPD
Sbjct: 302 YRTRDEISGVRQERDPVERIRSLILAHNIATEAELKDIEKENRKVVDEAIAKAKESPMPD 361
Query: 366 PSDLFTNVYVKGLGVEACGADRKEVKATLP 395
PS+LFTNVYVKG GVEA GADRKE++ATLP
Sbjct: 362 PSELFTNVYVKGFGVEAYGADRKELRATLP 391
>ODPA_ARATH (P52901) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 389
Score = 624 bits (1610), Expect = e-178
Identities = 302/395 (76%), Positives = 347/395 (87%), Gaps = 6/395 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR SS + + +P+ SA R IS+ +T +T+ETS+PFT+H C+PPS +V++
Sbjct: 1 MALSRLSSRS--NIITRPF---SAAFSRLISTDTTP-ITIETSLPFTAHLCDPPSRSVES 54
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
++ EL+ FF M LMRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEAAI +KD +IT
Sbjct: 55 SSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKDAIIT 114
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHC FL RGG+L EVFSELMGR+ GCSKGKGGSMHFY+KE FYGGHGIVGAQ+PLG
Sbjct: 115 AYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLG 174
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
G+AF QKYNK+ VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTAEW
Sbjct: 175 CGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEW 234
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
R+AKSP+YYKRGDY PGLKVDGMD AVKQACKFAK+HAL+ GP+ILEMDTYRYHGHSMS
Sbjct: 235 RAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGPIILEMDTYRYHGHSMS 294
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIER++KLVL+HD++TEKELKD+EKE RK+VD+AIAKAK+
Sbjct: 295 DPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRKEVDDAIAKAKD 354
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
MP+PS+LFTNVYVKG G E+ G DRKEVKA+LP
Sbjct: 355 CPMPEPSELFTNVYVKGFGTESFGPDRKEVKASLP 389
>ODPA_YEAST (P16387) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 420
Score = 371 bits (952), Expect = e-102
Identities = 190/345 (55%), Positives = 244/345 (70%), Gaps = 8/345 (2%)
Query: 39 TVETSIP---FTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFC 95
TV+ +P F S+ EPP + +T+ + L+ + DMV++RRME+A D+LYKAK IRGFC
Sbjct: 52 TVQIELPESSFESYMLEPPDLSYETSKATLLQMYKDMVIIRRMEMACDALYKAKKIRGFC 111
Query: 96 HLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGG 155
HL GQEA+AVG+E AI + D +IT+YR H RG ++ V +ELMGR+ G S GKGG
Sbjct: 112 HLSVGQEAIAVGIENAITKLDSIITSYRCHGFTFMRGASVKAVLAELMGRRAGVSYGKGG 171
Query: 156 SMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALN 215
SMH Y GFYGG+GIVGAQ+PLG GLAF +Y + +FTLYGDGA+NQGQ+FE+ N
Sbjct: 172 SMHLYAP--GFYGGNGIVGAQVPLGAGLAFAHQYKNEDACSFTLYGDGASNQGQVFESFN 229
Query: 216 IAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFA 275
+A LW+LP + CENN YGMGTA RS+ Y+KRG Y PGLKV+GMD+LAV QA KFA
Sbjct: 230 MAKLWNLPVVFCCENNKYGMGTAASRSSAMTEYFKRGQYIPGLKVNGMDILAVYQASKFA 289
Query: 276 KEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDI 334
K+ L GPL+LE +TYRY GHSMSDPG+TYRTRDEI +R + DPI ++ ++ I
Sbjct: 290 KDWCLSGKGPLVLEYETYRYGGHSMSDPGTTYRTRDEIQHMRSKNDPIAGLKMHLIDLGI 349
Query: 335 STEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
+TE E+K +K ARK VDE + A + P+ S LF +VYVKG
Sbjct: 350 ATEAEVKAYDKSARKYVDEQVELADAAPPPEAKLSILFEDVYVKG 394
>ODPA_SCHPO (Q10489) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 409
Score = 369 bits (947), Expect = e-102
Identities = 186/335 (55%), Positives = 234/335 (69%), Gaps = 5/335 (1%)
Query: 46 FTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVA 105
F + + PST ++ T EL+ + MV +RR+E+A D+LYKAK IRGFCHL GQEAVA
Sbjct: 59 FEGYKIDVPSTEIEVTKGELLGLYEKMVTIRRLELACDALYKAKKIRGFCHLSIGQEAVA 118
Query: 106 VGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGG 165
G+E AI D +IT+YR H RG ++ + ELMGR+ G SKGKGGSMH + K
Sbjct: 119 AGIEGAITLDDSIITSYRCHGFAYTRGLSIRSIIGELMGRQCGASKGKGGSMHIFAKN-- 176
Query: 166 FYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAI 225
FYGG+GIVGAQIPLG G+ F QKY + P TF LYGDGA+NQGQ FEA N+A LW LP I
Sbjct: 177 FYGGNGIVGAQIPLGAGIGFAQKYLEKPTTTFALYGDGASNQGQAFEAFNMAKLWGLPVI 236
Query: 226 LVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNG-P 284
CENN YGMGT+ RS+ +YKRG Y PGL V+GMDVLAV QA KFAK++ ++N P
Sbjct: 237 FACENNKYGMGTSAERSSAMTEFYKRGQYIPGLLVNGMDVLAVLQASKFAKKYTVENSQP 296
Query: 285 LILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIE 344
L++E TYRY GHSMSDPG+TYR+R+E+ VR RDPIE ++K ++ ++ ELK+IE
Sbjct: 297 LLMEFVTYRYGGHSMSDPGTTYRSREEVQKVRAARDPIEGLKKHIMEWGVANANELKNIE 356
Query: 345 KEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
K R VDE + A+ES PDP LF++VYV G
Sbjct: 357 KRIRGMVDEEVRIAEESPFPDPIEESLFSDVYVAG 391
>ODPA_KLULA (O13366) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
Length = 412
Score = 365 bits (936), Expect = e-100
Identities = 192/361 (53%), Positives = 247/361 (68%), Gaps = 13/361 (3%)
Query: 28 RSISSSSTETLT-----VETSIPFTSHN---CEPPSTTVQTTASELMSFFNDMVLMRRME 79
R++S + ET V+ +P TS + P + QTT S L+ + DM+++RRME
Sbjct: 28 RALSQVADETKPGDDDLVQIDLPETSFEGYLLDVPELSYQTTKSNLLQMYKDMIIVRRME 87
Query: 80 IAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVF 139
+A D+LYKAK IRGFCH GQEA+AVG+E AI ++D VIT+YR H RG + V
Sbjct: 88 MACDALYKAKKIRGFCHSSVGQEAIAVGIENAITKRDTVITSYRCHGFTYMRGAAVQAVL 147
Query: 140 SELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTL 199
+ELMGR+ G S GKGGSMH Y GFYGG+GIVGAQ+PLG GLAF +Y + +F L
Sbjct: 148 AELMGRRTGVSFGKGGSMHLYAP--GFYGGNGIVGAQVPLGAGLAFAHQYKHEDACSFAL 205
Query: 200 YGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLK 259
YGDGA+NQGQ+FE+ N+A LW+LPA+ CENN YGMGTA RS+ Y+KRG Y PGLK
Sbjct: 206 YGDGASNQGQVFESFNMAKLWNLPAVFCCENNKYGMGTAAARSSAMTEYFKRGQYIPGLK 265
Query: 260 VDGMDVLAVKQACKFAKEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQE 318
V+GMD+LAV QA KFAK+ + NGP++LE +TYRY GHSMSDPG+TYRTRDEI +R +
Sbjct: 266 VNGMDILAVYQASKFAKDWTVSGNGPIVLEYETYRYGGHSMSDPGTTYRTRDEIQHMRSK 325
Query: 319 RDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVK 376
DPI ++ +L I+TE E+K +K ARK VDE + A + P+ S LF +VYV
Sbjct: 326 NDPIAGLKMHLLELGIATEDEIKAYDKAARKYVDEQVELADAAPAPEAKMSILFEDVYVP 385
Query: 377 G 377
G
Sbjct: 386 G 386
>ODPA_CAEEL (P52899) Probable pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)
Length = 397
Score = 365 bits (936), Expect = e-100
Identities = 194/366 (53%), Positives = 246/366 (67%), Gaps = 6/366 (1%)
Query: 22 SSAIRHRSISSSSTETLTVETSIPFTSHNCEP-PSTTVQTTASELMSFFNDMVLMRRMEI 80
+S IR + + +STE ++ T P H + P+T+V + + ++ DM ++RRME
Sbjct: 13 ASGIRTQQVRLASTE-VSFHTK-PCKLHKLDNGPNTSVTLNREDALKYYRDMQVIRRMES 70
Query: 81 AADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFS 140
AA +LYK K IRGFCHLY GQEA AVGM+AA+ D VITAYR H G T+ EV +
Sbjct: 71 AAGNLYKEKKIRGFCHLYSGQEACAVGMKAAMTEGDAVITAYRCHGWTWLLGATVTEVLA 130
Query: 141 ELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLY 200
EL GR G GKGGSMH Y K FYGG+GIVGAQ PLG G+A KY + NV TLY
Sbjct: 131 ELTGRVAGNVHGKGGSMHMYTKN--FYGGNGIVGAQQPLGAGVALAMKYREQKNVCVTLY 188
Query: 201 GDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKV 260
GDGAANQGQLFEA N+A LWDLP + VCENN +GMGT RS+ S YY RGDY PG+ V
Sbjct: 189 GDGAANQGQLFEATNMAKLWDLPVLFVCENNGFGMGTTAERSSASTEYYTRGDYVPGIWV 248
Query: 261 DGMDVLAVKQACKFAKEHA-LKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQER 319
DGMD+LAV++A K+AKE+ GPL++EM TYRYHGHSMSDPG++YRTR+EI VR+ R
Sbjct: 249 DGMDILAVREATKWAKEYCDSGKGPLMMEMATYRYHGHSMSDPGTSYRTREEIQEVRKTR 308
Query: 320 DPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLG 379
DPI + ++ ++TE+ELK I+KE RK+VDEA+ A + P L+ ++Y
Sbjct: 309 DPITGFKDRIITSSLATEEELKAIDKEVRKEVDEALKIATSDGVLPPEALYADIYHNTPA 368
Query: 380 VEACGA 385
E GA
Sbjct: 369 QEIRGA 374
>ODPA_SMIMA (P52900) Pyruvate dehydrogenase E1 component alpha
subunit, mitochondrial precursor (EC 1.2.4.1) (PDHE1-A)
(Fragment)
Length = 363
Score = 358 bits (920), Expect = 1e-98
Identities = 178/337 (52%), Positives = 235/337 (68%), Gaps = 3/337 (0%)
Query: 52 EPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAA 111
E P TT T E + ++ M +RRME+ AD LYK K+IRGFCHLYDGQEA +G+EA
Sbjct: 21 EGPPTTAVLTREEGLKYYKIMQTVRRMELKADQLYKQKIIRGFCHLYDGQEACCMGLEAG 80
Query: 112 INRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHG 171
IN D VITAYR H RG + E+ +EL GR+ GC+KGKGGSMH Y K FYGG+G
Sbjct: 81 INPTDHVITAYRAHGFTYTRGLPVREILAELTGRRGGCAKGKGGSMHMYAKN--FYGGNG 138
Query: 172 IVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENN 231
IVGAQ+PLGVG+A KYN+ + TLYGDGAANQGQ+FEA N+AALW LP I +CENN
Sbjct: 139 IVGAQVPLGVGIALACKYNEKDEICLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENN 198
Query: 232 HYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMD 290
YGMGT+ R+A S YYKRGD+ PG+ VDGMDVL V++A KFA + GP+++E+
Sbjct: 199 RYGMGTSVERAAASTDYYKRGDFIPGIMVDGMDVLCVREATKFAAAYCRSGKGPMLMELQ 258
Query: 291 TYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQ 350
TYRYHGHSMSDPG +YRTR+EI VR + DPI ++ ++ +++++ +ELK+I+ E RK+
Sbjct: 259 TYRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNNNLASIEELKEIDVEVRKE 318
Query: 351 VDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
+++A A P +L ++Y + E GA++
Sbjct: 319 IEDAAQFATADPEPPLEELGYHIYSRDPPFEVRGANQ 355
>ODPA_HUMAN (P08559) Pyruvate dehydrogenase E1 component alpha
subunit, somatic form, mitochondrial precursor (EC
1.2.4.1) (PDHE1-A type I)
Length = 390
Score = 352 bits (903), Expect = 9e-97
Identities = 178/336 (52%), Positives = 232/336 (68%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP TTV T L ++ M +RRME+ AD LYK K+IRGFCHL DGQEA VG+EA I
Sbjct: 50 PPVTTVLTREDGL-KYYRMMQTVRRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGI 108
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D +ITAYR H RG ++ E+ +EL GRK GC+KGKGGSMH Y K FYGG+GI
Sbjct: 109 NPTDHLITAYRAHGFTFTRGLSVREILAELTGRKGGCAKGKGGSMHMYAKN--FYGGNGI 166
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+A KYN V TLYGDGAANQGQ+FEA N+AALW LP I +CENN
Sbjct: 167 VGAQVPLGAGIALACKYNGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENNR 226
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ R+A S YYKRGD+ PGL+VDGMD+L V++A +FA + GP+++E+ T
Sbjct: 227 YGMGTSVERAAASTDYYKRGDFIPGLRVDGMDILCVREATRFAAAYCRSGKGPILMELQT 286
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+EI VR + DPI ++ ++ ++++ +ELK+I+ E RK++
Sbjct: 287 YRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNSNLASVEELKEIDVEVRKEI 346
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
++A A P +L ++Y E GA++
Sbjct: 347 EDAAQFATADPEPPLEELGYHIYSSDPPFEVRGANQ 382
>ODPT_RAT (Q06437) Pyruvate dehydrogenase E1 component alpha
subunit, testis-specific form, mitochondrial precursor
(EC 1.2.4.1) (PDHE1-A type II)
Length = 391
Score = 351 bits (901), Expect = 2e-96
Identities = 173/336 (51%), Positives = 236/336 (69%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP++TV T E + ++ +M ++RRME+ AD LYK K IRGFCHL DGQEA VG+EA I
Sbjct: 51 PPTSTV-LTREEALKYYRNMQVIRRMELKADQLYKQKFIRGFCHLCDGQEACNVGLEAGI 109
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D +IT+YR H RG ++ + +EL GRK GC+KGKGGSMH Y K FYGG+GI
Sbjct: 110 NPTDHIITSYRAHGLCYTRGLSVKSILAELTGRKGGCAKGKGGSMHMYAKN--FYGGNGI 167
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+A KY K+ + LYGDGAANQGQ+FEA N++ALW LP + +CENN
Sbjct: 168 VGAQVPLGAGVALACKYLKNGQICLALYGDGAANQGQVFEAYNMSALWKLPCVFICENNR 227
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGTA RSA S Y+K+G PGL+V+GMD+L+V++A KFA +H GP+++E+ T
Sbjct: 228 YGMGTAIERSAASTDYHKKGFVIPGLRVNGMDILSVREATKFAADHCRSGKGPIVMELQT 287
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+E+ VR + DPI +R+ ++++++S+ +ELK+I+ + +K+V
Sbjct: 288 YRYHGHSMSDPGISYRTREEVQNVRSKSDPIMLLRERMISNNLSSVEELKEIDADVKKEV 347
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
+EA A P DL +Y + E GA +
Sbjct: 348 EEAAQFATTDPEPPLEDLANYLYHQNPPFEVRGAHK 383
>ODPT_HUMAN (P29803) Pyruvate dehydrogenase E1 component alpha
subunit, testis-specific form, mitochondrial precursor
(EC 1.2.4.1) (PDHE1-A type II)
Length = 388
Score = 351 bits (901), Expect = 2e-96
Identities = 179/335 (53%), Positives = 230/335 (68%), Gaps = 4/335 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP TTV T A E + ++ M+ +RRME+ AD LYK K IRGFCHL DGQEA VG+EA I
Sbjct: 48 PPVTTVLTRA-EGLKYYRMMLTVRRMELKADQLYKQKFIRGFCHLCDGQEACCVGLEAGI 106
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D VIT+YR H RG ++ + +EL GR+ GC+KGKGGSMH Y K FYGG+GI
Sbjct: 107 NPSDHVITSYRAHGVCYTRGLSVRSILAELTGRRGGCAKGKGGSMHMYTKN--FYGGNGI 164
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ PLG G+A KY + + TLYGDGAANQGQ+ EA N+AALW LP + +CENN
Sbjct: 165 VGAQGPLGAGIALACKYKGNDEICLTLYGDGAANQGQIAEAFNMAALWKLPCVFICENNL 224
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ R+A SP YYKRG++ PGLKVDGMDVL V++A KFA + GP+++E+ T
Sbjct: 225 YGMGTSTERAAASPDYYKRGNFIPGLKVDGMDVLCVREATKFAANYCRSGKGPILMELQT 284
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+EI VR +RDPI ++ ++ ++T +ELK+I E RK++
Sbjct: 285 YRYHGHSMSDPGVSYRTREEIQEVRSKRDPIIILQDRMVNSKLATVEELKEIGAEVRKEI 344
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGAD 386
D+A A P +L ++Y E GA+
Sbjct: 345 DDAAQFATTDPEPHLEELGHHIYSSDSSFEVRGAN 379
>ODPA_PIG (P29804) Pyruvate dehydrogenase E1 component alpha
subunit, somatic form, mitochondrial precursor (EC
1.2.4.1) (PDHE1-A type I) (Fragment)
Length = 389
Score = 350 bits (899), Expect = 3e-96
Identities = 176/336 (52%), Positives = 231/336 (68%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP TTV T L ++ M +RRME+ AD LYK K+IRGFCHL DGQEA VG+EA I
Sbjct: 49 PPVTTVLTREDGL-KYYRMMQTVRRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGI 107
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D +ITAYR H RG ++ E+ +EL GR+ GC KGKGGSMH Y K FYGG+GI
Sbjct: 108 NPTDHLITAYRAHGFTFTRGLSVREILAELTGRRGGCGKGKGGSMHMYAKN--FYGGNGI 165
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+A KYN V TLYGDGAANQGQ+FEA N+AALW LP + +CENN
Sbjct: 166 VGAQVPLGAGIALACKYNGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCVFICENNR 225
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ R+A S YYKRGD+ PGL+VDGMD+L V++A +FA + GP+++E+ T
Sbjct: 226 YGMGTSVERAAASTDYYKRGDFIPGLRVDGMDILCVREATRFAAAYCRSGKGPILMELQT 285
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+EI VR + DPI ++ ++ ++++ +ELK+I+ E RK++
Sbjct: 286 YRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNSNLASVEELKEIDVEVRKEI 345
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
++A A P +L ++Y E GA++
Sbjct: 346 EDAAQFATADPEPPLEELGYHIYCNDPPFEVRGANQ 381
>ODPA_RAT (P26284) Pyruvate dehydrogenase E1 component alpha
subunit, somatic form, mitochondrial precursor (EC
1.2.4.1) (PDHE1-A type I)
Length = 390
Score = 350 bits (897), Expect = 5e-96
Identities = 178/336 (52%), Positives = 230/336 (67%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP TTV T L ++ M +RRME+ AD LYK K+IRGFCHL DGQEA VG+EA I
Sbjct: 50 PPVTTVLTREDGL-KYYRMMQTVRRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGI 108
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D +ITAYR H RG + V +EL GR+ GC+KGKGGSMH Y K FYGG+GI
Sbjct: 109 NPTDHLITAYRAHGFTFNRGHAVRAVLAELTGRRGGCAKGKGGSMHMYAKN--FYGGNGI 166
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+A KYN V TLYGDGAANQGQ+FEA N+AALW LP I +CENN
Sbjct: 167 VGAQVPLGAGIALACKYNGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENNR 226
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ R+A S YYKRGD+ PGL+VDGMD+L V++A KFA + GP+++E+ T
Sbjct: 227 YGMGTSVERAAASTDYYKRGDFIPGLRVDGMDILCVREATKFAAAYCRSGKGPILMELQT 286
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+EI VR + DPI ++ ++ ++++ +ELK+I+ E RK++
Sbjct: 287 YRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNSNLASVEELKEIDVEVRKEI 346
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
++A A P +L ++Y E GA++
Sbjct: 347 EDAAQFATADPEPPLEELGYHIYSSDPPFEVRGANQ 382
>ODPT_MOUSE (P35487) Pyruvate dehydrogenase E1 component alpha
subunit, testis-specific form, mitochondrial precursor
(EC 1.2.4.1) (PDHE1-A type II)
Length = 391
Score = 349 bits (895), Expect = 8e-96
Identities = 173/336 (51%), Positives = 235/336 (69%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP++TV T A E + ++ M ++RRME+ AD LYK K IRGFCHL DGQEA VG+EA I
Sbjct: 51 PPTSTVLTRA-EALKYYRTMQVIRRMELKADQLYKQKFIRGFCHLCDGQEACCVGLEAGI 109
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D VIT+YR H RG ++ + +EL GRK GC+KGKGGSMH Y K FYGG+GI
Sbjct: 110 NPTDHVITSYRAHGFCYTRGLSVKSILAELTGRKGGCAKGKGGSMHMYGKN--FYGGNGI 167
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+AF KY K+ V LYGDGAANQGQ+FEA N++ALW LP + +CENN
Sbjct: 168 VGAQVPLGAGVAFACKYLKNGQVCLALYGDGAANQGQVFEAYNMSALWKLPCVFICENNL 227
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ RSA S Y+K+G PGL+V+GMD+L V++A KFA +H GP+++E+ T
Sbjct: 228 YGMGTSNERSAASTDYHKKGFIIPGLRVNGMDILCVREATKFAADHCRSGKGPIVMELQT 287
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YR+R+E+ VR + DPI +R+ ++++++S +ELK+I+ + +K+V
Sbjct: 288 YRYHGHSMSDPGISYRSREEVHNVRSKSDPIMLLRERIISNNLSNIEELKEIDADVKKEV 347
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
++A A P D+ +Y + E GA +
Sbjct: 348 EDAAQFATTDPEPAVEDIANYLYHQDPPFEVRGAHK 383
>ODPA_MOUSE (P35486) Pyruvate dehydrogenase E1 component alpha
subunit, somatic form, mitochondrial precursor (EC
1.2.4.1) (PDHE1-A type I)
Length = 390
Score = 349 bits (895), Expect = 8e-96
Identities = 177/336 (52%), Positives = 230/336 (67%), Gaps = 4/336 (1%)
Query: 53 PPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI 112
PP TTV T L ++ M +RRME+ AD LYK K+IRGFCHL DGQEA VG+EA I
Sbjct: 50 PPVTTVLTREDGL-KYYRMMQTVRRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGI 108
Query: 113 NRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGI 172
N D +ITAYR H RG + + +EL GR+ GC+KGKGGSMH Y K FYGG+GI
Sbjct: 109 NPTDHLITAYRAHGFTFTRGLPVRAILAELTGRRGGCAKGKGGSMHMYAKN--FYGGNGI 166
Query: 173 VGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNH 232
VGAQ+PLG G+A KYN V TLYGDGAANQGQ+FEA N+AALW LP I +CENN
Sbjct: 167 VGAQVPLGAGIALACKYNGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENNR 226
Query: 233 YGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALK-NGPLILEMDT 291
YGMGT+ R+A S YYKRGD+ PGL+VDGMD+L V++A KFA + GP+++E+ T
Sbjct: 227 YGMGTSVERAAASTDYYKRGDFIPGLRVDGMDILCVREATKFAAAYCRSGKGPILMELQT 286
Query: 292 YRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQV 351
YRYHGHSMSDPG +YRTR+EI VR + DPI ++ ++ ++++ +ELK+I+ E RK++
Sbjct: 287 YRYHGHSMSDPGVSYRTREEIQEVRSKSDPIMLLKDRMVNSNLASVEELKEIDVEVRKEI 346
Query: 352 DEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADR 387
++A A P +L ++Y E GA++
Sbjct: 347 EDAAQFATADPEPPLEELGYHIYSSDPPFEVRGANQ 382
>ODPA_ASCSU (P26267) Pyruvate dehydrogenase E1 component alpha
subunit type I, mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)
Length = 396
Score = 345 bits (886), Expect = 9e-95
Identities = 179/364 (49%), Positives = 241/364 (66%), Gaps = 6/364 (1%)
Query: 13 STLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEP-PSTTVQTTASELMSFFND 71
+ + K +S ++ S+ +STE T +T PF H + P V T + + ++
Sbjct: 6 ANIFKVPTVSPSVMAISVRLASTEA-TFQTK-PFKLHKLDSGPDINVHVTKEDAVHYYTQ 63
Query: 72 MVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCR 131
M+ +RRME AA +LYK K +RGFCHLY GQEA AVG +AA++ D +TAYR H
Sbjct: 64 MLTIRRMESAAGNLYKEKKVRGFCHLYSGQEACAVGTKAAMDAGDAAVTAYRCHGWTYLS 123
Query: 132 GGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNK 191
G ++ +V EL GR G GKGGSMH Y + FYGG+GIVGAQ PLG G+AF KY K
Sbjct: 124 GSSVAKVLCELTGRITGNVYGKGGSMHMYGEN--FYGGNGIVGAQQPLGTGIAFAMKYRK 181
Query: 192 DPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKR 251
+ NV T++GDGA NQGQLFE++N+A LWDLP + VCENN YGMGTA RS+ S YY R
Sbjct: 182 EKNVCITMFGDGATNQGQLFESMNMAKLWDLPVLYVCENNGYGMGTAAARSSASTDYYTR 241
Query: 252 GDYAPGLKVDGMDVLAVKQACKFAKEHA-LKNGPLILEMDTYRYHGHSMSDPGSTYRTRD 310
GDY PG+ VDGMDVLAV+QA ++AKE GPL++EM TYRY GHSMSDPG++YRTR+
Sbjct: 242 GDYVPGIWVDGMDVLAVRQAVRWAKEWCNAGKGPLMIEMATYRYSGHSMSDPGTSYRTRE 301
Query: 311 EISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLF 370
E+ VR+ RDPI + ++ + TE E+K+I+K+ RK++D A+ +A + +
Sbjct: 302 EVQEVRKTRDPITGFKDKIVTAGLVTEDEIKEIDKQVRKEIDAAVKQAHTDKESPVELML 361
Query: 371 TNVY 374
T++Y
Sbjct: 362 TDIY 365
>ODPT_ASCSU (P26268) Pyruvate dehydrogenase E1 component alpha
subunit type II, mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A) (Fragment)
Length = 391
Score = 340 bits (872), Expect = 4e-93
Identities = 181/364 (49%), Positives = 234/364 (63%), Gaps = 6/364 (1%)
Query: 13 STLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEP-PSTTVQTTASELMSFFND 71
S + K + S++ S +STE T + PF H + P + T + + ++
Sbjct: 1 SNIFKGPTVGSSVVAMSARLASTEA-TFQAK-PFKLHKLDSGPDVNMHVTKEDALRYYTQ 58
Query: 72 MVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCR 131
M +RRME AA +LYK K +RGFCHLY GQEA AVGM+AA+ D ITAYR H
Sbjct: 59 MQTIRRMETAAGNLYKEKKVRGFCHLYSGQEACAVGMKAAMEPGDAAITAYRCHGWTYLS 118
Query: 132 GGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNK 191
G + +V EL GR G GKGGSMH Y + FYGG+GIVGAQ PLG G+AF KY K
Sbjct: 119 GSPVAKVLCELTGRITGNVYGKGGSMHMYGEN--FYGGNGIVGAQQPLGTGIAFAMKYKK 176
Query: 192 DPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKR 251
NV TL+GDGA NQGQL+E++N+A LW+LP + VCENN YGMGT+ RS+ S YY R
Sbjct: 177 QKNVCITLFGDGATNQGQLYESMNMAKLWELPVLYVCENNGYGMGTSAARSSASTDYYTR 236
Query: 252 GDYAPGLKVDGMDVLAVKQACKFAKEHA-LKNGPLILEMDTYRYHGHSMSDPGSTYRTRD 310
GDY PG VDGMDVLAV+QA ++ KE GPL++EM TYRY GHSMSDPG++YRTR+
Sbjct: 237 GDYVPGFWVDGMDVLAVRQAIRWGKEWCNAGKGPLMIEMATYRYGGHSMSDPGTSYRTRE 296
Query: 311 EISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLF 370
EI VR+ RDPI + ++ + TE ELK+++KE RK+VD A+ +A + L
Sbjct: 297 EIQEVRKTRDPITGFKDKIVTAGLVTEDELKEVDKEIRKEVDAAVKQAHTDKEAPVEMLL 356
Query: 371 TNVY 374
T++Y
Sbjct: 357 TDIY 360
>ODPA_RHIME (Q9R9N5) Pyruvate dehydrogenase E1 component, alpha
subunit (EC 1.2.4.1)
Length = 348
Score = 298 bits (764), Expect = 1e-80
Identities = 165/349 (47%), Positives = 215/349 (61%), Gaps = 10/349 (2%)
Query: 28 RSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYK 87
+S S SS +T F T + + + + + +M+L+RR E A LY
Sbjct: 5 KSASVSSRKTAAKPAKKDFAG------GTIAEFSKEDDLKAYREMLLIRRFEEKAGQLYG 58
Query: 88 AKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKD 147
I GFCHLY GQEAV VGM+ A+ D VIT YRDH L G + V +EL GR+
Sbjct: 59 MGFIGGFCHLYIGQEAVVVGMQLALKEGDQVITGYRDHGHMLACGMSARGVMAELTGRRG 118
Query: 148 GCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQ 207
G SKGKGGSMH + KE FYGGHGIVGAQ+ LG GLAF +Y + NV+ +GDGAANQ
Sbjct: 119 GLSKGKGGSMHMFSKEKHFYGGHGIVGAQVSLGTGLAFANRYRGNDNVSLAYFGDGAANQ 178
Query: 208 GQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDY--APGLKVDGMDV 265
GQ++E+ N+AALW LP I + ENN Y MGT+ R++ + +RG PG +VDGMDV
Sbjct: 179 GQVYESFNMAALWKLPVIYIVENNRYAMGTSVSRASAQTDFSQRGASFGIPGYQVDGMDV 238
Query: 266 LAVKQACKFAKEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIER 324
AVK A A EH GP+ILEM TYRY GHSMSDP + YR++DE+ +R E DPIE+
Sbjct: 239 RAVKAAADEAVEHCRSGKGPIILEMLTYRYRGHSMSDP-AKYRSKDEVQKMRSEHDPIEQ 297
Query: 325 VRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNV 373
V+ + +TE ELK I+KE R V ++ A+ PD S+L+T++
Sbjct: 298 VKARLTDKGWATEDELKQIDKEVRDIVADSADFAQSDPEPDVSELYTDI 346
>ODPA_RICCN (Q92IS3) Pyruvate dehydrogenase E1 component, alpha
subunit (EC 1.2.4.1)
Length = 326
Score = 286 bits (731), Expect = 8e-77
Identities = 151/317 (47%), Positives = 198/317 (61%), Gaps = 5/317 (1%)
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
T E + F DM+L+RR E LY I GFCHLY GQEAV ++ + D IT
Sbjct: 11 TKEEYIKSFKDMLLLRRFEEKCGQLYGMGEIGGFCHLYIGQEAVISAIDMVKQKGDSTIT 70
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
+YRDH + G V +ELMGR GCSKGKGGSMH + FYGGHGIVGAQ+P+G
Sbjct: 71 SYRDHAHIILAGTEPKYVLAELMGRATGCSKGKGGSMHLFNVPNKFYGGHGIVGAQVPIG 130
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
GLAF +KYN N+ FT GDGA NQGQ++EA N+AALW LP + + ENN Y MGT+
Sbjct: 131 TGLAFVEKYNDTHNICFTFLGDGAVNQGQVYEAFNMAALWGLPVVYIIENNEYSMGTSVA 190
Query: 241 RSAKSPAYYKRGDY--APGLKVDGMDVLAVKQACKFAKEHALKNG-PLILEMDTYRYHGH 297
RS YK+G G ++DGMD + K A E+ +N PLILE+ TYRY GH
Sbjct: 191 RSTFMRDLYKKGASFGIKGFQLDGMDFEEMYDGSKQAAEYVRENSFPLILEVKTYRYRGH 250
Query: 298 SMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAK 357
SMSDP + YR+++E+ + ERDP+ +RK +L + TE +LK IE+ ++ V EA+
Sbjct: 251 SMSDP-AKYRSKEEVEQYK-ERDPLVIIRKTILDNKYVTEADLKAIEQSVKEIVKEAVEF 308
Query: 358 AKESQMPDPSDLFTNVY 374
++ S +PD +L+T VY
Sbjct: 309 SENSPLPDEGELYTQVY 325
>ODPA_ZYMMO (O66112) Pyruvate dehydrogenase E1 component, alpha
subunit (EC 1.2.4.1)
Length = 354
Score = 279 bits (714), Expect = 8e-75
Identities = 156/340 (45%), Positives = 213/340 (61%), Gaps = 8/340 (2%)
Query: 40 VETSIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYD 99
V ++IP +H+ P EL+ F+ M+++RR E LY LI GFCHLY
Sbjct: 16 VGSAIP--NHDLPPIPGRYHADREELLEFYRRMLMIRRFEERCGQLYGLGLIAGFCHLYI 73
Query: 100 GQEAVAVGMEAAINR-KDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMH 158
GQEAVAVG++AA+ +D VIT YR+H L G V +EL GR G S GKGGSMH
Sbjct: 74 GQEAVAVGLQAALQPGRDSVITGYREHGHMLAYGIDPKIVMAELTGRASGISHGKGGSMH 133
Query: 159 FYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAA 218
+ E F+GG+GIVGAQ+PLG GLAF KY D + +GDG+ANQGQ++EA N+AA
Sbjct: 134 MFSTEHKFFGGNGIVGAQVPLGAGLAFAHKYRNDGGCSAAYFGDGSANQGQVYEAYNMAA 193
Query: 219 LWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGD--YAPGLKVDGMDVLAVKQACKFAK 276
LW LP I V ENN Y MGT+ R+ A +RG P L VDGMDVL V+ A A
Sbjct: 194 LWKLPVIFVIENNGYAMGTSIQRANAHTALSERGAGFGIPALVVDGMDVLEVRGAATVAV 253
Query: 277 EHA-LKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDIS 335
+ GP+ILEM TYRY GHSMSDP + YR+R+E++ +++ DP++ ++K + A +
Sbjct: 254 DWVQAGKGPIILEMKTYRYRGHSMSDP-ARYRSREEVNDMKENHDPLDNLKKDLFAAGV- 311
Query: 336 TEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYV 375
E EL ++++ R+QV EA A+++ +P +L+TN+ V
Sbjct: 312 PEAELVKLDEDIRQQVKEAADFAEKAPLPADEELYTNILV 351
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,350,920
Number of Sequences: 164201
Number of extensions: 2103538
Number of successful extensions: 6186
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 6029
Number of HSP's gapped (non-prelim): 132
length of query: 395
length of database: 59,974,054
effective HSP length: 112
effective length of query: 283
effective length of database: 41,583,542
effective search space: 11768142386
effective search space used: 11768142386
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146588.4


