

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.12 - phase: 0 /pseudo
(840 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
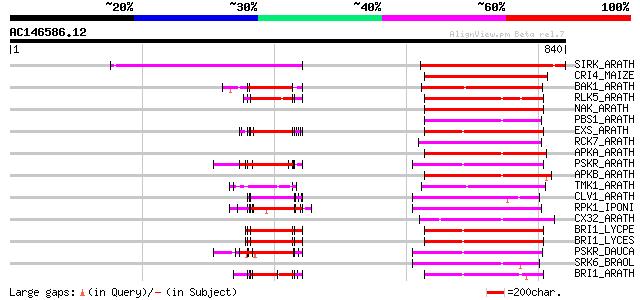
Score E
Sequences producing significant alignments: (bits) Value
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 231 6e-60
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 152 3e-36
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 142 3e-33
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 142 5e-33
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 141 8e-33
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 139 2e-32
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 139 4e-32
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 138 5e-32
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 134 1e-30
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 131 8e-30
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 130 2e-29
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 129 3e-29
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 128 5e-29
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 128 7e-29
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 126 2e-28
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 123 2e-27
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 123 2e-27
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 119 4e-26
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 117 1e-25
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 117 1e-25
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 231 bits (589), Expect = 6e-60
Identities = 126/292 (43%), Positives = 173/292 (59%), Gaps = 3/292 (1%)
Query: 153 TMKLTSHLQYKDDVYDRMWFSYELI-DWRRLSTSLNNDHLVQNIYKPPTIVMSTAATPVN 211
T KL S +YKDD+YDR+W + +++ L+TSL D + N Y+P + VMSTA T N
Sbjct: 205 TGKLPS--RYKDDIYDRIWTPRIVSSEYKILNTSLTVDQFLNNGYQPASTVMSTAETARN 262
Query: 212 ASAPLQFHWSSNNENDQYYLYIHFNEVEELAANETREFNITVNDKLWFGPVTPIYRTPDL 271
S L + + N ++Y+Y+HF E+E L +N+TREF+I +N+ + Y D
Sbjct: 263 ESLYLTLSFRPPDPNAKFYVYMHFAEIEVLKSNQTREFSIWLNEDVISPSFKLRYLLTDT 322
Query: 272 IFSTEPLRRAETYQISLSKTKNSTLPPILNAFEIYMAKDFSQLETQQDDVDNITNIKNAY 331
+ +P+ L LPPI+NA E+Y +F Q+ T DVD + IK Y
Sbjct: 323 FVTPDPVSGITINFSLLQPPGEFVLPPIINALEVYQVNEFLQIPTHPQDVDAMRKIKATY 382
Query: 332 GVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQY 391
V +NWQGDPC PV+Y WEG++C D+ PR+ SLN+S S L G+I + S LT ++
Sbjct: 383 RVKKNWQGDPCVPVDYSWEGIDCIQSDNTTNPRVVSLNISFSELRGQIDPAFSNLTSIRK 442
Query: 392 LDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
LDLS N+L G +P FL L +L LNV N LTG+VP L ERSK GSLSLR
Sbjct: 443 LDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLHERSKNGSLSLR 494
Score = 207 bits (528), Expect = 7e-53
Identities = 100/219 (45%), Positives = 152/219 (68%), Gaps = 2/219 (0%)
Query: 622 AVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGL 681
A + S +L+W ERL I++DAA GL+YLHNGCKPP +HRD+KP+NILL+E + AK+ADFGL
Sbjct: 659 AGKRSFILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGL 718
Query: 682 SRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRAS 741
SR+F + IST AG+ GY+DP++ T N+K+D+YS G+VLLE+ITG+ A+ +
Sbjct: 719 SRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSK 778
Query: 742 GESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQI 801
E +HI V I+ GDIR I+D RL+ ++D+ SAWK+ EIA++ T +RP MSQ+
Sbjct: 779 TEKVHISDHVRSILANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQV 838
Query: 802 LAELKECLSLDMVHRNNGRERAIVELTSLNIASDTIPLA 840
+ ELK+ + + + N + ++ ++N+ ++ +P A
Sbjct: 839 VMELKQIVYGIVTDQENYDDS--TKMLTVNLDTEMVPRA 875
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 152 bits (385), Expect = 3e-36
Identities = 76/185 (41%), Positives = 116/185 (62%), Gaps = 1/185 (0%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
LNW R+ IAV AA G++YLH PP +HRD+K SNIL+DE+ +A++ADFGLS D
Sbjct: 603 LNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPAD 662
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHIL 748
+ +S PAGT GY+DP++ R K+D+YSFG+VLLE+++G+KA + E +I+
Sbjct: 663 SGTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKA-IDMQFEEGNIV 721
Query: 749 QWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELKEC 808
+W P+++ GDI +I+D L D+ + K+ +A +RP M ++ L+
Sbjct: 722 EWAVPLIKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHA 781
Query: 809 LSLDM 813
L+L M
Sbjct: 782 LALLM 786
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 142 bits (359), Expect = 3e-33
Identities = 68/186 (36%), Positives = 120/186 (63%), Gaps = 4/186 (2%)
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
E+ L+W +R IA+ +A GL YLH+ C P +HRD+K +NILLDE A + DFGL++
Sbjct: 380 ESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAK 439
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA--LVR-A 740
D D+H++T GT G++ P++ TG +++K D++ +G++LLELITG++A L R A
Sbjct: 440 LMDYK-DTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLA 498
Query: 741 SGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQ 800
+ + + +L WV +++ + +++D LQG + +++++A+ T +ERP MS+
Sbjct: 499 NDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSE 558
Query: 801 ILAELK 806
++ L+
Sbjct: 559 VVRMLE 564
Score = 57.0 bits (136), Expect = 2e-07
Identities = 36/125 (28%), Positives = 66/125 (52%), Gaps = 11/125 (8%)
Query: 322 DNITNIKNAYG----VTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTG 377
D ++ +KN+ V ++W P W + C++D+ +T ++L ++ L+G
Sbjct: 30 DALSALKNSLADPNKVLQSWDATLVTPCT--WFHVTCNSDNS-----VTRVDLGNANLSG 82
Query: 378 EISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKT 437
++ + +L LQYL+L +N++ G +P+ L L L L++ NNL+G +PS L K
Sbjct: 83 QLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGRLKKL 142
Query: 438 GSLSL 442
L L
Sbjct: 143 RFLRL 147
Score = 54.7 bits (130), Expect = 1e-06
Identities = 28/68 (41%), Positives = 45/68 (66%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + SL+L + L+G I S++ +L L++L L+NNSL+G +P L + +LQVL++
Sbjct: 114 NLTELVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSN 173
Query: 421 NNLTGLVP 428
N LTG +P
Sbjct: 174 NPLTGDIP 181
Score = 53.5 bits (127), Expect = 2e-06
Identities = 31/81 (38%), Positives = 47/81 (57%), Gaps = 5/81 (6%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P + L L S+ +TG I + LT L LDL N+L+GP+P L +L+ L+ L + N+
Sbjct: 92 PNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNNS 151
Query: 423 LTGLVPSELLERSKTGSLSLR 443
L+G +P RS T L+L+
Sbjct: 152 LSGEIP-----RSLTAVLTLQ 167
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 142 bits (357), Expect = 5e-33
Identities = 75/182 (41%), Positives = 112/182 (61%), Gaps = 4/182 (2%)
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRA--F 685
+L W ERL IA+DAA GL YLH+ C PP +HRD+K SNILLD + AK+ADFG+++
Sbjct: 787 VLGWPERLRIALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQM 846
Query: 686 DNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESI 745
+ AG+ GY+ P++ T N+K+DIYSFG+VLLEL+TGK+ G+
Sbjct: 847 SGSKTPEAMSGIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDK- 905
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
+ +WV +++ + +ID +L KF KV+ I + TSP+ + RP M +++ L
Sbjct: 906 DMAKWVCTALDKCGLEPVIDPKLDLKFK-EEISKVIHIGLLCTSPLPLNRPSMRKVVIML 964
Query: 806 KE 807
+E
Sbjct: 965 QE 966
Score = 51.6 bits (122), Expect = 9e-06
Identities = 27/64 (42%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ LNL+++ L+GEI + L +L YLDLS+N +G +P L L+ L VLN+ N+L+
Sbjct: 525 LNELNLANNHLSGEIPKEVGILPVLNYLDLSSNQFSGEIPLELQNLK-LNVLNLSYNHLS 583
Query: 425 GLVP 428
G +P
Sbjct: 584 GKIP 587
Score = 48.5 bits (114), Expect = 7e-05
Identities = 23/67 (34%), Positives = 40/67 (59%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+T + LS++ L+G+I L L L+LS+NS G +P +I ++L L + KN +
Sbjct: 405 LTRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFS 464
Query: 425 GLVPSEL 431
G +P+E+
Sbjct: 465 GSIPNEI 471
Score = 48.5 bits (114), Expect = 7e-05
Identities = 25/67 (37%), Positives = 40/67 (59%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
I ++ + + +GEI S+ KL L LDLS N L+G +P L ++L LN+ N+L+
Sbjct: 477 IIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLSGEIPRELRGWKNLNELNLANNHLS 536
Query: 425 GLVPSEL 431
G +P E+
Sbjct: 537 GEIPKEV 543
Score = 48.1 bits (113), Expect = 9e-05
Identities = 23/67 (34%), Positives = 42/67 (62%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +L+L+ + LTG I S I++L ++ ++L NNS +G LP+ + + +L+ + N LT
Sbjct: 238 LVNLDLTFNQLTGSIPSWITQLKTVEQIELFNNSFSGELPESMGNMTTLKRFDASMNKLT 297
Query: 425 GLVPSEL 431
G +P L
Sbjct: 298 GKIPDNL 304
Score = 47.8 bits (112), Expect = 1e-04
Identities = 27/78 (34%), Positives = 45/78 (57%), Gaps = 1/78 (1%)
Query: 355 STDDDNNPPRITSLNLSSSGLTGEISSSIS-KLTMLQYLDLSNNSLNGPLPDFLIQLRSL 413
S DD + + SL+LS + L G I S+ L L++L++S N+L+ +P + R L
Sbjct: 106 SADDFDTCHNLISLDLSENLLVGSIPKSLPFNLPNLKFLEISGNNLSDTIPSSFGEFRKL 165
Query: 414 QVLNVGKNNLTGLVPSEL 431
+ LN+ N L+G +P+ L
Sbjct: 166 ESLNLAGNFLSGTIPASL 183
Score = 46.2 bits (108), Expect = 4e-04
Identities = 23/71 (32%), Positives = 41/71 (57%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + L L+ L G I S+S+LT L LDL+ N L G +P ++ QL++++ + +
Sbjct: 210 NLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKTVEQIELFN 269
Query: 421 NNLTGLVPSEL 431
N+ +G +P +
Sbjct: 270 NSFSGELPESM 280
Score = 45.1 bits (105), Expect = 8e-04
Identities = 22/78 (28%), Positives = 43/78 (54%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+++L +S + +G I + I L + + + N +G +P+ L++L+ L L++ KN L+
Sbjct: 453 LSNLRISKNRFSGSIPNEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLS 512
Query: 425 GLVPSELLERSKTGSLSL 442
G +P EL L+L
Sbjct: 513 GEIPRELRGWKNLNELNL 530
Score = 43.9 bits (102), Expect = 0.002
Identities = 23/79 (29%), Positives = 41/79 (51%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L L + +GEIS+++ K L + LSNN L+G +P L L +L + N+
Sbjct: 380 KLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSF 439
Query: 424 TGLVPSELLERSKTGSLSL 442
TG +P ++ +L +
Sbjct: 440 TGSIPKTIIGAKNLSNLRI 458
Score = 42.0 bits (97), Expect = 0.007
Identities = 22/80 (27%), Positives = 41/80 (50%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
PR++ L LS + TG I +I L L +S N +G +P+ + L + ++ +N+
Sbjct: 427 PRLSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFSGSIPNEIGSLNGIIEISGAEND 486
Query: 423 LTGLVPSELLERSKTGSLSL 442
+G +P L++ + L L
Sbjct: 487 FSGEIPESLVKLKQLSRLDL 506
Score = 42.0 bits (97), Expect = 0.007
Identities = 28/85 (32%), Positives = 41/85 (47%)
Query: 358 DDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLN 417
D+ N + SLNL + L G + SI++ L L L NN L G LP L LQ ++
Sbjct: 302 DNLNLLNLESLNLFENMLEGPLPESITRSKTLSELKLFNNRLTGVLPSQLGANSPLQYVD 361
Query: 418 VGKNNLTGLVPSELLERSKTGSLSL 442
+ N +G +P+ + K L L
Sbjct: 362 LSYNRFSGEIPANVCGEGKLEYLIL 386
Score = 41.2 bits (95), Expect = 0.012
Identities = 28/84 (33%), Positives = 41/84 (48%), Gaps = 6/84 (7%)
Query: 349 WEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPL-PDFL 407
W G++C + + S++LSS L G S + L L L L NNS+NG L D
Sbjct: 56 WLGVSCDATSN-----VVSVDLSSFMLVGPFPSILCHLPSLHSLSLYNNSINGSLSADDF 110
Query: 408 IQLRSLQVLNVGKNNLTGLVPSEL 431
+L L++ +N L G +P L
Sbjct: 111 DTCHNLISLDLSENLLVGSIPKSL 134
Score = 41.2 bits (95), Expect = 0.012
Identities = 23/68 (33%), Positives = 39/68 (56%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
+++ L+LS + L+GEI + L L+L+NN L+G +P + L L L++ N
Sbjct: 500 QLSRLDLSKNQLSGEIPRELRGWKNLNELNLANNHLSGEIPKEVGILPVLNYLDLSSNQF 559
Query: 424 TGLVPSEL 431
+G +P EL
Sbjct: 560 SGEIPLEL 567
Score = 40.8 bits (94), Expect = 0.015
Identities = 29/82 (35%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query: 363 PRITSLNLSSSGLTGEISSS-ISKLTMLQYLDLSNNSLNGPLPDFL-IQLRSLQVLNVGK 420
P + SL+L ++ + G +S+ L LDLS N L G +P L L +L+ L +
Sbjct: 89 PSLHSLSLYNNSINGSLSADDFDTCHNLISLDLSENLLVGSIPKSLPFNLPNLKFLEISG 148
Query: 421 NNLTGLVPSELLERSKTGSLSL 442
NNL+ +PS E K SL+L
Sbjct: 149 NNLSDTIPSSFGEFRKLESLNL 170
Score = 40.4 bits (93), Expect = 0.020
Identities = 24/65 (36%), Positives = 36/65 (54%)
Query: 378 EISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKT 437
+I S + LT LQ L L+ +L GP+P L +L SL L++ N LTG +PS + +
Sbjct: 203 QIPSQLGNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKTV 262
Query: 438 GSLSL 442
+ L
Sbjct: 263 EQIEL 267
Score = 40.0 bits (92), Expect = 0.026
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLN-GPLPDFLIQLRSLQVLNVGKNN 422
++ SLNL+ + L+G I +S+ +T L+ L L+ N + +P L L LQVL + N
Sbjct: 164 KLESLNLAGNFLSGTIPASLGNVTTLKELKLAYNLFSPSQIPSQLGNLTELQVLWLAGCN 223
Query: 423 LTGLVPSELLERSKTGSLSL 442
L G +P L + +L L
Sbjct: 224 LVGPIPPSLSRLTSLVNLDL 243
Score = 38.1 bits (87), Expect = 0.098
Identities = 25/84 (29%), Positives = 45/84 (52%), Gaps = 11/84 (13%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
++LS + +GEI +++ L+YL L +NS +G + + L + +SL + + N L+G +
Sbjct: 360 VDLSYNRFSGEIPANVCGEGKLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQI 419
Query: 428 PS-----------ELLERSKTGSL 440
P EL + S TGS+
Sbjct: 420 PHGFWGLPRLSLLELSDNSFTGSI 443
Score = 35.8 bits (81), Expect = 0.48
Identities = 20/67 (29%), Positives = 37/67 (54%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ L L ++ LTG + S + + LQY+DLS N +G +P + L+ L + N+ +
Sbjct: 333 LSELKLFNNRLTGVLPSQLGANSPLQYVDLSYNRFSGEIPANVCGEGKLEYLILIDNSFS 392
Query: 425 GLVPSEL 431
G + + L
Sbjct: 393 GEISNNL 399
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 141 bits (355), Expect = 8e-33
Identities = 72/181 (39%), Positives = 117/181 (63%), Gaps = 3/181 (1%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+WN R+ +A+ AA GL +LHN +P ++RD K SNILLD N +AK++DFGL+R
Sbjct: 173 LSWNTRVRMALGAARGLAFLHNA-QPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMG 231
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIH-I 747
+SH+STR GT GY P++ TG+ + K+D+YSFG+VLLEL++G++A+ + H +
Sbjct: 232 DNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNL 291
Query: 748 LQWVTP-IVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAELK 806
+ W P + + + ++D RLQG++ + A K+ +A+ S RP M++I+ ++
Sbjct: 292 VDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTME 351
Query: 807 E 807
E
Sbjct: 352 E 352
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 139 bits (351), Expect = 2e-32
Identities = 71/180 (39%), Positives = 108/180 (59%), Gaps = 4/180 (2%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+WN R+ IA AA GL++LH+ PP ++RD K SNILLDE H K++DFGL++
Sbjct: 182 LDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTG 241
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
SH+STR GT+GY P++ TG K+D+YSFG+V LELITG+KA+ GE +
Sbjct: 242 DKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQ-N 300
Query: 747 ILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
++ W P+ +R + D RL+G+F + ++ + +A RP ++ ++ L
Sbjct: 301 LVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 139 bits (349), Expect = 4e-32
Identities = 74/182 (40%), Positives = 112/182 (60%), Gaps = 3/182 (1%)
Query: 628 MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDN 687
+L+W++RL IAV AA GL +LH+G P +HRD+K SNILLD + K+ADFGL+R +
Sbjct: 1011 VLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLI-S 1069
Query: 688 DIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVR--ASGESI 745
+SH+ST AGTFGY+ P++ ++ K D+YSFG++LLEL+TGK+ E
Sbjct: 1070 ACESHVSTVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGG 1129
Query: 746 HILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
+++ W + +G +ID L NS ++++IAM + +RP+M +L L
Sbjct: 1130 NLVGWAIQKINQGKAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
Query: 806 KE 807
KE
Sbjct: 1190 KE 1191
Score = 60.8 bits (146), Expect = 1e-08
Identities = 29/67 (43%), Positives = 43/67 (63%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L + + TGEI S + LT L+YLD+S N L+G +P + L +L+ LN+ KNNL
Sbjct: 725 KLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSENLLSGEIPTKICGLPNLEFLNLAKNNL 784
Query: 424 TGLVPSE 430
G VPS+
Sbjct: 785 RGEVPSD 791
Score = 56.2 bits (134), Expect = 3e-07
Identities = 32/81 (39%), Positives = 49/81 (59%), Gaps = 1/81 (1%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPL-PDFLIQLRSLQVLNVG 419
N + +L+LS + LTG + +S+L L YLDLS+N +G L P F I L +L L+V
Sbjct: 111 NLKHLQTLDLSGNSLTGLLPRLLSELPQLLYLDLSDNHFSGSLPPSFFISLPALSSLDVS 170
Query: 420 KNNLTGLVPSELLERSKTGSL 440
N+L+G +P E+ + S +L
Sbjct: 171 NNSLSGEIPPEIGKLSNLSNL 191
Score = 54.3 bits (129), Expect = 1e-06
Identities = 32/85 (37%), Positives = 46/85 (53%), Gaps = 6/85 (7%)
Query: 349 WEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLI 408
W G+ C R+ SL+L S L G+I IS L L+ L L+ N +G +P +
Sbjct: 57 WVGVTCLLG------RVNSLSLPSLSLRGQIPKEISSLKNLRELCLAGNQFSGKIPPEIW 110
Query: 409 QLRSLQVLNVGKNNLTGLVPSELLE 433
L+ LQ L++ N+LTGL+P L E
Sbjct: 111 NLKHLQTLDLSGNSLTGLLPRLLSE 135
Score = 53.5 bits (127), Expect = 2e-06
Identities = 27/64 (42%), Positives = 41/64 (63%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ ++LS++ L+GEI +S+S+LT L LDLS N+L G +P + LQ LN+ N L
Sbjct: 606 LVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALTGSIPKEMGNSLKLQGLNLANNQLN 665
Query: 425 GLVP 428
G +P
Sbjct: 666 GHIP 669
Score = 53.5 bits (127), Expect = 2e-06
Identities = 30/67 (44%), Positives = 40/67 (58%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+T L+LS + LTG I + LQ L+L+NN LNG +P+ L SL LN+ KN L
Sbjct: 630 LTILDLSGNALTGSIPKEMGNSLKLQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLD 689
Query: 425 GLVPSEL 431
G VP+ L
Sbjct: 690 GPVPASL 696
Score = 52.0 bits (123), Expect = 7e-06
Identities = 29/71 (40%), Positives = 39/71 (54%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + L LS + LTGEI I KLT L L+L+ N G +P L SL L++G
Sbjct: 470 NAASLKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVELGDCTSLTTLDLGS 529
Query: 421 NNLTGLVPSEL 431
NNL G +P ++
Sbjct: 530 NNLQGQIPDKI 540
Score = 49.7 bits (117), Expect = 3e-05
Identities = 25/66 (37%), Positives = 40/66 (59%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ LNL+++ G+I + T L LDL +N+L G +PD + L LQ L + NNL+
Sbjct: 498 LSVLNLNANMFQGKIPVELGDCTSLTTLDLGSNNLQGQIPDKITALAQLQCLVLSYNNLS 557
Query: 425 GLVPSE 430
G +PS+
Sbjct: 558 GSIPSK 563
Score = 48.9 bits (115), Expect = 6e-05
Identities = 28/76 (36%), Positives = 43/76 (55%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N ++ LNL+++ L G I S L L L+L+ N L+GP+P L L+ L +++
Sbjct: 650 NSLKLQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVPASLGNLKELTHMDLSF 709
Query: 421 NNLTGLVPSELLERSK 436
NNL+G + SEL K
Sbjct: 710 NNLSGELSSELSTMEK 725
Score = 47.8 bits (112), Expect = 1e-04
Identities = 25/67 (37%), Positives = 39/67 (57%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ LNL+ + L G + +S+ L L ++DLS N+L+G L L + L L + +N T
Sbjct: 678 LVKLNLTKNKLDGPVPASLGNLKELTHMDLSFNNLSGELSSELSTMEKLVGLYIEQNKFT 737
Query: 425 GLVPSEL 431
G +PSEL
Sbjct: 738 GEIPSEL 744
Score = 47.8 bits (112), Expect = 1e-04
Identities = 26/74 (35%), Positives = 43/74 (57%)
Query: 369 NLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
+LS + L+G I + + +L + LSNN L+G +P L +L +L +L++ N LTG +P
Sbjct: 586 DLSYNRLSGPIPEELGECLVLVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALTGSIP 645
Query: 429 SELLERSKTGSLSL 442
E+ K L+L
Sbjct: 646 KEMGNSLKLQGLNL 659
Score = 47.0 bits (110), Expect = 2e-04
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query: 363 PRITSLNLSSSGLTGEISSSIS-KLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKN 421
P++ L+LS + +G + S L L LD+SNNSL+G +P + +L +L L +G N
Sbjct: 137 PQLLYLDLSDNHFSGSLPPSFFISLPALSSLDVSNNSLSGEIPPEIGKLSNLSNLYMGLN 196
Query: 422 NLTGLVPSEL 431
+ +G +PSE+
Sbjct: 197 SFSGQIPSEI 206
Score = 45.1 bits (105), Expect = 8e-04
Identities = 31/91 (34%), Positives = 46/91 (50%)
Query: 352 LNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLR 411
+N S +D + +L+L S+ TGEI S+ K T L S N L G LP +
Sbjct: 413 INGSIPEDLWKLPLMALDLDSNNFTGEIPKSLWKSTNLMEFTASYNRLEGYLPAEIGNAA 472
Query: 412 SLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
SL+ L + N LTG +P E+ + + L+L
Sbjct: 473 SLKRLVLSDNQLTGEIPREIGKLTSLSVLNL 503
Score = 43.5 bits (101), Expect = 0.002
Identities = 25/64 (39%), Positives = 36/64 (56%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ L L+ + +G+I I L LQ LDLS NSL G LP L +L L L++ N+ +
Sbjct: 91 LRELCLAGNQFSGKIPPEIWNLKHLQTLDLSGNSLTGLLPRLLSELPQLLYLDLSDNHFS 150
Query: 425 GLVP 428
G +P
Sbjct: 151 GSLP 154
Score = 41.6 bits (96), Expect = 0.009
Identities = 26/66 (39%), Positives = 33/66 (49%)
Query: 377 GEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSK 436
G + ISKL L LDLS N L +P +L +L +LN+ L GL+P EL
Sbjct: 224 GPLPKEISKLKHLAKLDLSYNPLKCSIPKSFGELHNLSILNLVSAELIGLIPPELGNCKS 283
Query: 437 TGSLSL 442
SL L
Sbjct: 284 LKSLML 289
Score = 40.8 bits (94), Expect = 0.015
Identities = 25/78 (32%), Positives = 42/78 (53%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ SL L+++ +GEI I ML++L L++N L+G +P L SL+ +++ N L+
Sbjct: 331 LDSLLLANNRFSGEIPHEIEDCPMLKHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLS 390
Query: 425 GLVPSELLERSKTGSLSL 442
G + S G L L
Sbjct: 391 GTIEEVFDGCSSLGELLL 408
Score = 40.8 bits (94), Expect = 0.015
Identities = 25/72 (34%), Positives = 38/72 (52%)
Query: 371 SSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSE 430
S + L G + + I L+ L LS+N L G +P + +L SL VLN+ N G +P E
Sbjct: 456 SYNRLEGYLPAEIGNAASLKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVE 515
Query: 431 LLERSKTGSLSL 442
L + + +L L
Sbjct: 516 LGDCTSLTTLDL 527
Score = 40.4 bits (93), Expect = 0.020
Identities = 23/67 (34%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +++LS + L+G I + L L L+NN +NG +P+ L +L L L++ NN T
Sbjct: 379 LEAIDLSGNLLSGTIEEVFDGCSSLGELLLTNNQINGSIPEDLWKL-PLMALDLDSNNFT 437
Query: 425 GLVPSEL 431
G +P L
Sbjct: 438 GEIPKSL 444
Score = 38.1 bits (87), Expect = 0.098
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ LNL S+ L G I + L+ L LS NSL+GPLP L ++ L + +N L+
Sbjct: 260 LSILNLVSAELIGLIPPELGNCKSLKSLMLSFNSLSGPLPLELSEI-PLLTFSAERNQLS 318
Query: 425 GLVPS 429
G +PS
Sbjct: 319 GSLPS 323
Score = 37.4 bits (85), Expect = 0.17
Identities = 29/104 (27%), Positives = 45/104 (42%), Gaps = 24/104 (23%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSL----------------------- 399
P ++SL++S++ L+GEI I KL+ L L + NS
Sbjct: 162 PALSSLDVSNNSLSGEIPPEIGKLSNLSNLYMGLNSFSGQIPSEIGNISLLKNFAAPSCF 221
Query: 400 -NGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
NGPLP + +L+ L L++ N L +P E L+L
Sbjct: 222 FNGPLPKEISKLKHLAKLDLSYNPLKCSIPKSFGELHNLSILNL 265
Score = 37.4 bits (85), Expect = 0.17
Identities = 21/69 (30%), Positives = 36/69 (51%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P + L+L+S+ L+G I + L+ +DLS N L+G + + SL L + N
Sbjct: 353 PMLKHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLSGTIEEVFDGCSSLGELLLTNNQ 412
Query: 423 LTGLVPSEL 431
+ G +P +L
Sbjct: 413 INGSIPEDL 421
Score = 37.0 bits (84), Expect = 0.22
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 1/82 (1%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + SL LS + L+G + +S++ +L + N L+G LP ++ + + L L +
Sbjct: 280 NCKSLKSLMLSFNSLSGPLPLELSEIPLLTF-SAERNQLSGSLPSWMGKWKVLDSLLLAN 338
Query: 421 NNLTGLVPSELLERSKTGSLSL 442
N +G +P E+ + LSL
Sbjct: 339 NRFSGEIPHEIEDCPMLKHLSL 360
Score = 35.0 bits (79), Expect = 0.83
Identities = 19/57 (33%), Positives = 32/57 (55%)
Query: 375 LTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSEL 431
L+G + S + K +L L L+NN +G +P + L+ L++ N L+G +P EL
Sbjct: 317 LSGSLPSWMGKWKVLDSLLLANNRFSGEIPHEIEDCPMLKHLSLASNLLSGSIPREL 373
Score = 34.3 bits (77), Expect = 1.4
Identities = 21/67 (31%), Positives = 36/67 (53%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N ++ L++S + L+GEI + I L L++L+L+ N+L G +P + + L G
Sbjct: 746 NLTQLEYLDVSENLLSGEIPTKICGLPNLEFLNLAKNNLRGEVPSDGVCQDPSKALLSGN 805
Query: 421 NNLTGLV 427
L G V
Sbjct: 806 KELCGRV 812
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 138 bits (348), Expect = 5e-32
Identities = 72/190 (37%), Positives = 115/190 (59%), Gaps = 3/190 (1%)
Query: 619 YLHAVENSNM-LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIA 677
+LH + + L+WN R+ IA AA GL+YLH+ PP ++RDLK SNILL E+ K++
Sbjct: 188 HLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLS 247
Query: 678 DFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL 737
DFGL++ + +H+STR GT+GY P + TG K+DIYSFG+VLLELITG+KA+
Sbjct: 248 DFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAI 307
Query: 738 VRA-SGESIHILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVER 795
+ + +++ W P+ +R + ++D LQG++ + ++ + I+ R
Sbjct: 308 DNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMR 367
Query: 796 PDMSQILAEL 805
P +S ++ L
Sbjct: 368 PVVSDVVLAL 377
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 134 bits (336), Expect = 1e-30
Identities = 74/187 (39%), Positives = 120/187 (63%), Gaps = 5/187 (2%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W RL +A+ AA GL +LH+ + ++RD K SNILLD +AK++DFGL++
Sbjct: 173 LSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIG 231
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
SH+STR GT GY P++ TG+ K+D+YSFG+VLLEL++G++A+ R SGE +
Sbjct: 232 DKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGER-N 290
Query: 747 ILQWVTP-IVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
+++W P +V + I +ID RLQ ++ + A KV +++ + RP+MS++++ L
Sbjct: 291 LVEWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHL 350
Query: 806 KECLSLD 812
+ SL+
Sbjct: 351 EHIQSLN 357
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 131 bits (329), Expect = 8e-30
Identities = 71/200 (35%), Positives = 111/200 (55%), Gaps = 3/200 (1%)
Query: 610 HFQNFETNAYLHAVENSN-MLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILL 668
+ +N + +LH + +L W RL IA AA GL YLH GC P +HRD+K SNILL
Sbjct: 809 YMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILL 868
Query: 669 DENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLL 728
DEN ++ +ADFGL+R + ++H+ST GT GY+ P++ + K D+YSFG+VLL
Sbjct: 869 DENFNSHLADFGLARLM-SPYETHVSTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLL 927
Query: 729 ELITGKKALVRASGESIH-ILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSS 787
EL+T K+ + + ++ WV + + D + K + ++V+EIA
Sbjct: 928 ELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFRVLEIACLC 987
Query: 788 TSPIEVERPDMSQILAELKE 807
S +RP Q+++ L +
Sbjct: 988 LSENPKQRPTTQQLVSWLDD 1007
Score = 58.5 bits (140), Expect = 7e-08
Identities = 27/83 (32%), Positives = 51/83 (60%), Gaps = 3/83 (3%)
Query: 349 WEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLI 408
W G+ C++ NN R+ L L + L+G++S S+ KL ++ L+LS N + +P +
Sbjct: 65 WTGITCNS---NNTGRVIRLELGNKKLSGKLSESLGKLDEIRVLNLSRNFIKDSIPLSIF 121
Query: 409 QLRSLQVLNVGKNNLTGLVPSEL 431
L++LQ L++ N+L+G +P+ +
Sbjct: 122 NLKNLQTLDLSSNDLSGGIPTSI 144
Score = 56.6 bits (135), Expect = 3e-07
Identities = 30/73 (41%), Positives = 46/73 (62%)
Query: 357 DDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVL 416
++ N ++ +L + L+G I SS+S +T L+ LDLSNN L+G +P L QL L
Sbjct: 541 EEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKF 600
Query: 417 NVGKNNLTGLVPS 429
+V NNL+G++PS
Sbjct: 601 SVAYNNLSGVIPS 613
Score = 48.9 bits (115), Expect = 6e-05
Identities = 35/135 (25%), Positives = 64/135 (46%), Gaps = 3/135 (2%)
Query: 309 KDFSQLETQQDDVDNITNIKNAYGVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSL 368
K+F L ++ NI +A G+ ++ + + + G D + ++ L
Sbjct: 361 KNFESLSYFSLSNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPDDSSLHFEKLKVL 420
Query: 369 NLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
+++ LTG + +S LQ LDLS N L G +P ++ ++L L++ N+ TG +P
Sbjct: 421 VVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIP 480
Query: 429 SELLERSKTGSLSLR 443
L +K SL+ R
Sbjct: 481 KSL---TKLESLTSR 492
Score = 46.6 bits (109), Expect = 3e-04
Identities = 23/68 (33%), Positives = 37/68 (53%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N P + LNL ++ L+G + + + + L LDL N NG LP+ L + L+ +N+ +
Sbjct: 290 NSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLAR 349
Query: 421 NNLTGLVP 428
N G VP
Sbjct: 350 NTFHGQVP 357
Score = 45.1 bits (105), Expect = 8e-04
Identities = 26/54 (48%), Positives = 31/54 (57%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKN 421
L+LS + LTG I S I L YLDLSNNS G +P L +L SL N+ N
Sbjct: 444 LDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTSRNISVN 497
Score = 43.1 bits (100), Expect = 0.003
Identities = 23/76 (30%), Positives = 38/76 (49%)
Query: 367 SLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGL 426
++ L + L+G I L L DL N+L+G +P L + SL+ L++ N L+G
Sbjct: 527 TIELGHNNLSGPIWEEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGS 586
Query: 427 VPSELLERSKTGSLSL 442
+P L + S S+
Sbjct: 587 IPVSLQQLSFLSKFSV 602
Score = 43.1 bits (100), Expect = 0.003
Identities = 21/72 (29%), Positives = 39/72 (54%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N +I + L+ + G +S K +L++L L N L G +P+ L L+ L +L +
Sbjct: 169 HNSTQIRVVKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQ 228
Query: 420 KNNLTGLVPSEL 431
+N L+G + E+
Sbjct: 229 ENRLSGSLSREI 240
Score = 41.6 bits (96), Expect = 0.009
Identities = 29/106 (27%), Positives = 48/106 (44%), Gaps = 4/106 (3%)
Query: 342 CAPVNYMWEGLNCST----DDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN 397
C + ++ G+N T +D + R+ L + + L+G +S I L+ L LD+S N
Sbjct: 195 CVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVSWN 254
Query: 398 SLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
+G +PD +L L+ N G +P L L+LR
Sbjct: 255 LFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLR 300
Score = 41.2 bits (95), Expect = 0.012
Identities = 33/108 (30%), Positives = 51/108 (46%), Gaps = 26/108 (24%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFL----IQLR----- 411
N + +L+LSS+ L+G I +SI+ L LQ DLS+N NG LP + Q+R
Sbjct: 122 NLKNLQTLDLSSNDLSGGIPTSIN-LPALQSFDLSSNKFNGSLPSHICHNSTQIRVVKLA 180
Query: 412 ----------------SLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
L+ L +G N+LTG +P +L + L ++
Sbjct: 181 VNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQ 228
Score = 36.2 bits (82), Expect = 0.37
Identities = 21/71 (29%), Positives = 37/71 (51%)
Query: 372 SSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSEL 431
++G G I S++ L L+L NNSL+G L + +L L++G N G +P L
Sbjct: 277 TNGFIGGIPKSLANSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENL 336
Query: 432 LERSKTGSLSL 442
+ + +++L
Sbjct: 337 PDCKRLKNVNL 347
Score = 34.7 bits (78), Expect = 1.1
Identities = 27/78 (34%), Positives = 39/78 (49%), Gaps = 18/78 (23%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLI------QLRSLQ------- 414
L+LS++ TGEI S++KL + L N S+N P PDF R+LQ
Sbjct: 468 LDLSNNSFTGEIPKSLTKL---ESLTSRNISVNEPSPDFPFFMKRNESARALQYNQIFGF 524
Query: 415 --VLNVGKNNLTGLVPSE 430
+ +G NNL+G + E
Sbjct: 525 PPTIELGHNNLSGPIWEE 542
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 130 bits (326), Expect = 2e-29
Identities = 74/203 (36%), Positives = 123/203 (60%), Gaps = 13/203 (6%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
L+W RL +A+ AA GL +LHN + ++RD K SNILLD +AK++DFGL++
Sbjct: 174 LSWTLRLKVALGAAKGLAFLHNA-ETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTG 232
Query: 689 IDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKAL--VRASGESIH 746
SH+STR GT+GY P++ TG+ K+D+YS+G+VLLE+++G++A+ R GE
Sbjct: 233 DKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQ-K 291
Query: 747 ILQWVTPIV-ERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPDMSQILAEL 805
+++W P++ + + +ID RLQ ++ + A KV +A+ + RP+M+++++ L
Sbjct: 292 LVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHL 351
Query: 806 KECLSL--------DMVHRNNGR 820
+ +L DMV R R
Sbjct: 352 EHIQTLNEAGGRNIDMVQRRMRR 374
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 129 bits (324), Expect = 3e-29
Identities = 77/192 (40%), Positives = 108/192 (56%), Gaps = 5/192 (2%)
Query: 624 ENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSR 683
E L W +RL +A+D A G++YLH +HRDLKPSNILL ++M AK+ADFGL R
Sbjct: 681 EGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVR 740
Query: 684 AFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRAS-G 742
S I TR AGTFGY+ P++ TG K D+YSFG++L+ELITG+K+L +
Sbjct: 741 LAPEGKGS-IETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPE 799
Query: 743 ESIHILQWVTP--IVERGDIRSIIDARLQ-GKFDINSAWKVVEIAMSSTSPIEVERPDMS 799
ESIH++ W I + + ID + + + S V E+A + +RPDM
Sbjct: 800 ESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMG 859
Query: 800 QILAELKECLSL 811
+ L + L
Sbjct: 860 HAVNILSSLVEL 871
Score = 52.4 bits (124), Expect = 5e-06
Identities = 34/97 (35%), Positives = 48/97 (49%), Gaps = 11/97 (11%)
Query: 333 VTRNWQG-DPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQY 391
+ +W+G DPC W G+ CS + IT ++L LTG IS + LQ
Sbjct: 341 LAESWKGNDPCTN----WIGIACSNGN------ITVISLEKMELTGTISPEFGAIKSLQR 390
Query: 392 LDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
+ L N+L G +P L L +L+ L+V N L G VP
Sbjct: 391 IILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVP 427
Score = 49.3 bits (116), Expect = 4e-05
Identities = 31/94 (32%), Positives = 50/94 (52%), Gaps = 12/94 (12%)
Query: 340 DPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSL 399
DPC W + C+ R+T + + SGL G +S + L+ L+ L+L N++
Sbjct: 51 DPCK-----WTHIVCT-----GTKRVTRIQIGHSGLQGTLSPDLRNLSELERLELQWNNI 100
Query: 400 NGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLE 433
+GP+P L L SLQVL + NN +PS++ +
Sbjct: 101 SGPVPS-LSGLASLQVLMLSNNNFDS-IPSDVFQ 132
Score = 44.3 bits (103), Expect = 0.001
Identities = 27/69 (39%), Positives = 44/69 (63%), Gaps = 2/69 (2%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ SL L+ LTG+I+ + +T L+ + L +N +GPLPDF L+ L+ L++ N+
Sbjct: 210 QVQSLWLNGQKLTGDITV-LQNMTGLKEVWLHSNKFSGPLPDF-SGLKELESLSLRDNSF 267
Query: 424 TGLVPSELL 432
TG VP+ LL
Sbjct: 268 TGPVPASLL 276
Score = 41.6 bits (96), Expect = 0.009
Identities = 24/59 (40%), Positives = 35/59 (58%), Gaps = 1/59 (1%)
Query: 370 LSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
L S+ +G + S L L+ L L +NS GP+P L+ L SL+V+N+ N+L G VP
Sbjct: 239 LHSNKFSGPLPD-FSGLKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVP 296
Score = 36.6 bits (83), Expect = 0.28
Identities = 16/42 (38%), Positives = 27/42 (64%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDF 406
+ SL+L + TG + +S+ L L+ ++L+NN L GP+P F
Sbjct: 257 LESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVPVF 298
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 128 bits (322), Expect = 5e-29
Identities = 74/200 (37%), Positives = 112/200 (56%), Gaps = 10/200 (5%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ N LH + + L W R +AV+AA GL YLH+ C P +HRD+K +NILLD
Sbjct: 768 YMPNGSLGELLHGSKGGH-LQWETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLD 826
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
+ A +ADFGL++ + S + AG++GY+ P++ T ++K+D+YSFG+VLLE
Sbjct: 827 SDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLE 886
Query: 730 LITGKKALVRASGESIHILQWV-------TPIVERGDIRSIIDARLQGKFDINSAWKVVE 782
LI GKK V GE + I++WV T + + +I+D RL G + + S V +
Sbjct: 887 LIAGKKP-VGEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRLTG-YPLTSVIHVFK 944
Query: 783 IAMSSTSPIEVERPDMSQIL 802
IAM RP M +++
Sbjct: 945 IAMMCVEEEAAARPTMREVV 964
Score = 53.5 bits (127), Expect = 2e-06
Identities = 26/78 (33%), Positives = 47/78 (59%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ +N S++ +TG I SIS+ + L +DLS N +NG +P + +++L LN+ N LT
Sbjct: 506 LSRINTSANNITGGIPDSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNISGNQLT 565
Query: 425 GLVPSELLERSKTGSLSL 442
G +P+ + + +L L
Sbjct: 566 GSIPTGIGNMTSLTTLDL 583
Score = 50.8 bits (120), Expect = 1e-05
Identities = 28/77 (36%), Positives = 43/77 (55%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N + +L L + LTG I +S L L+ LDLS N L G +P I L ++ ++N+
Sbjct: 262 SNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTGEIPQSFINLGNITLINLF 321
Query: 420 KNNLTGLVPSELLERSK 436
+NNL G +P + E K
Sbjct: 322 RNNLYGQIPEAIGELPK 338
Score = 47.0 bits (110), Expect = 2e-04
Identities = 28/79 (35%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNN-SLNGPLP-DFLIQLRSLQVLNVGKNN 422
+ +L L+++ TGE+ + LT L+ L++SNN +L G P + L + L+VL+ NN
Sbjct: 96 LVNLTLAANNFTGELPLEMKSLTSLKVLNISNNGNLTGTFPGEILKAMVDLEVLDTYNNN 155
Query: 423 LTGLVPSELLERSKTGSLS 441
G +P E+ E K LS
Sbjct: 156 FNGKLPPEMSELKKLKYLS 174
Score = 46.6 bits (109), Expect = 3e-04
Identities = 27/69 (39%), Positives = 41/69 (59%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L+++S LTGEI +S+S L L L L N+L G +P L L SL+ L++ N L
Sbjct: 242 KLEILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQL 301
Query: 424 TGLVPSELL 432
TG +P +
Sbjct: 302 TGEIPQSFI 310
Score = 45.8 bits (107), Expect = 5e-04
Identities = 25/67 (37%), Positives = 40/67 (59%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ SL+LS + LTGEI S L + ++L N+L G +P+ + +L L+V V +NN T
Sbjct: 291 LKSLDLSINQLTGEIPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFT 350
Query: 425 GLVPSEL 431
+P+ L
Sbjct: 351 LQLPANL 357
Score = 44.7 bits (104), Expect = 0.001
Identities = 25/67 (37%), Positives = 39/67 (57%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ L++S + LTG I + + L+ L LSNN GP+P+ L + +SL + + KN L
Sbjct: 363 LIKLDVSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEELGKCKSLTKIRIVKNLLN 422
Query: 425 GLVPSEL 431
G VP+ L
Sbjct: 423 GTVPAGL 429
Score = 43.9 bits (102), Expect = 0.002
Identities = 28/82 (34%), Positives = 43/82 (52%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N IT +NL + L G+I +I +L L+ ++ N+ LP L + +L L+V
Sbjct: 311 NLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFTLQLPANLGRNGNLIKLDVSD 370
Query: 421 NNLTGLVPSELLERSKTGSLSL 442
N+LTGL+P +L K L L
Sbjct: 371 NHLTGLIPKDLCRGEKLEMLIL 392
Score = 43.9 bits (102), Expect = 0.002
Identities = 24/78 (30%), Positives = 39/78 (49%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ + LS++ +GEI +I LQ L L N G +P + +L+ L +N NN+T
Sbjct: 458 LDQIYLSNNWFSGEIPPAIGNFPNLQTLFLDRNRFRGNIPREIFELKHLSRINTSANNIT 517
Query: 425 GLVPSELLERSKTGSLSL 442
G +P + S S+ L
Sbjct: 518 GGIPDSISRCSTLISVDL 535
Score = 42.7 bits (99), Expect = 0.004
Identities = 25/63 (39%), Positives = 38/63 (59%), Gaps = 2/63 (3%)
Query: 368 LNLSSSG-LTGEISSSISKLTM-LQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTG 425
LN+S++G LTG I K + L+ LD NN+ NG LP + +L+ L+ L+ G N +G
Sbjct: 123 LNISNNGNLTGTFPGEILKAMVDLEVLDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSG 182
Query: 426 LVP 428
+P
Sbjct: 183 EIP 185
Score = 42.7 bits (99), Expect = 0.004
Identities = 25/67 (37%), Positives = 35/67 (51%)
Query: 376 TGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERS 435
TG + LT L+ LD+++ +L G +P L L+ L L + NNLTG +P EL
Sbjct: 230 TGGVPPEFGGLTKLEILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLV 289
Query: 436 KTGSLSL 442
SL L
Sbjct: 290 SLKSLDL 296
Score = 41.2 bits (95), Expect = 0.012
Identities = 23/68 (33%), Positives = 39/68 (56%), Gaps = 1/68 (1%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG-KNN 422
++ L+ + +GEI S + L+YL L+ L+G P FL +L++L+ + +G N+
Sbjct: 169 KLKYLSFGGNFFSGEIPESYGDIQSLEYLGLNGAGLSGKSPAFLSRLKNLREMYIGYYNS 228
Query: 423 LTGLVPSE 430
TG VP E
Sbjct: 229 YTGGVPPE 236
Score = 40.4 bits (93), Expect = 0.020
Identities = 20/45 (44%), Positives = 28/45 (61%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLP 404
NN + +LN+S + LTG I + I +T L LDLS N L+G +P
Sbjct: 549 NNVKNLGTLNISGNQLTGSIPTGIGNMTSLTTLDLSFNDLSGRVP 593
Score = 39.7 bits (91), Expect = 0.034
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N P +T + L+ + +GE+ ++S +L + LSNN +G +P + +LQ L + +
Sbjct: 431 NLPLVTIIELTDNFFSGELPVTMSG-DVLDQIYLSNNWFSGEIPPAIGNFPNLQTLFLDR 489
Query: 421 NNLTGLVPSELLE 433
N G +P E+ E
Sbjct: 490 NRFRGNIPREIFE 502
Score = 38.9 bits (89), Expect = 0.057
Identities = 20/82 (24%), Positives = 42/82 (50%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N P + +L L + G I I +L L ++ S N++ G +PD + + +L +++ +
Sbjct: 478 NFPNLQTLFLDRNRFRGNIPREIFELKHLSRINTSANNITGGIPDSISRCSTLISVDLSR 537
Query: 421 NNLTGLVPSELLERSKTGSLSL 442
N + G +P + G+L++
Sbjct: 538 NRINGEIPKGINNVKNLGTLNI 559
Score = 34.7 bits (78), Expect = 1.1
Identities = 17/64 (26%), Positives = 34/64 (52%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L+ ++ G++ +S+L L+YL N +G +P+ ++SL+ L + L+G
Sbjct: 149 LDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSGEIPESYGDIQSLEYLGLNGAGLSGKS 208
Query: 428 PSEL 431
P+ L
Sbjct: 209 PAFL 212
Score = 33.9 bits (76), Expect = 1.8
Identities = 20/65 (30%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSN-NSLNGPLPDFLIQLRSLQVLNVGKNNLTGL 426
L L+ +GL+G+ + +S+L L+ + + NS G +P L L++L++ LTG
Sbjct: 197 LGLNGAGLSGKSPAFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKLEILDMASCTLTGE 256
Query: 427 VPSEL 431
+P+ L
Sbjct: 257 IPTSL 261
Score = 33.1 bits (74), Expect = 3.1
Identities = 18/69 (26%), Positives = 36/69 (52%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P++ + + T ++ +++ + L LD+S+N L G +P L + L++L + N
Sbjct: 337 PKLEVFEVWENNFTLQLPANLGRNGNLIKLDVSDNHLTGLIPKDLCRGEKLEMLILSNNF 396
Query: 423 LTGLVPSEL 431
G +P EL
Sbjct: 397 FFGPIPEEL 405
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 128 bits (321), Expect = 7e-29
Identities = 75/201 (37%), Positives = 113/201 (55%), Gaps = 5/201 (2%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ +N + LH L+W+ R NIAV AHGL YLH C P +HRD+KP NILLD
Sbjct: 892 YMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLAYLHFDCDPAIVHRDIKPMNILLD 951
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
++ I+DFG+++ D S S GT GY+ P+ T ++++D+YS+G+VLLE
Sbjct: 952 SDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENAFTTVKSRESDVYSYGVVLLE 1011
Query: 730 LITGKKALVRASGESIHILQWVTPI-VERGDIRSIIDARLQGKFDINSAWKVVEIAMS-- 786
LIT KKAL + I+ WV + + G+I+ I+D L + +S + V A+S
Sbjct: 1012 LITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIVDPSLLDELIDSSVMEQVTEALSLA 1071
Query: 787 -STSPIEVE-RPDMSQILAEL 805
+ EV+ RP M ++ +L
Sbjct: 1072 LRCAEKEVDKRPTMRDVVKQL 1092
Score = 54.3 bits (129), Expect = 1e-06
Identities = 29/75 (38%), Positives = 47/75 (62%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L L + L GEI + L+ LQYL L N+L+G +P + +++SLQ L + +NNL+G +
Sbjct: 337 LQLQQNQLEGEIPGELGMLSQLQYLHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGEL 396
Query: 428 PSELLERSKTGSLSL 442
P ++ E + SL+L
Sbjct: 397 PVDMTELKQLVSLAL 411
Score = 52.8 bits (125), Expect = 4e-06
Identities = 33/99 (33%), Positives = 54/99 (54%), Gaps = 1/99 (1%)
Query: 345 VNYMWEGLNCSTDDD-NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPL 403
V + GLN S + N +T+L L + +G + SS+ +T LQ L L++N+L G L
Sbjct: 169 VYFTGNGLNGSIPSNIGNMSELTTLWLDDNQFSGPVPSSLGNITTLQELYLNDNNLVGTL 228
Query: 404 PDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
P L L +L L+V N+L G +P + + + ++SL
Sbjct: 229 PVTLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISL 267
Score = 52.4 bits (124), Expect = 5e-06
Identities = 29/79 (36%), Positives = 46/79 (57%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ LNLS + L G + S +S L LD S+N LNG +P L L L L++G+N+
Sbjct: 548 KLEHLNLSHNILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLTELTKLSLGENSF 607
Query: 424 TGLVPSELLERSKTGSLSL 442
+G +P+ L + +K +L L
Sbjct: 608 SGGIPTSLFQSNKLLNLQL 626
Score = 52.0 bits (123), Expect = 7e-06
Identities = 36/124 (29%), Positives = 59/124 (47%), Gaps = 31/124 (25%)
Query: 333 VTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKL------ 386
+T++W P + W G+ C + + +LNLSS G++GE IS L
Sbjct: 45 ITQSWNASDSTPCS--WLGVEC-----DRRQFVDTLNLSSYGISGEFGPEISHLKHLKKV 97
Query: 387 ------------------TMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
++L+++DLS+NS G +PD L L++L+ L++ N+L G P
Sbjct: 98 VLSGNGFFGSIPSQLGNCSLLEHIDLSSNSFTGNIPDTLGALQNLRNLSLFFNSLIGPFP 157
Query: 429 SELL 432
LL
Sbjct: 158 ESLL 161
Score = 51.2 bits (121), Expect = 1e-05
Identities = 30/82 (36%), Positives = 44/82 (53%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N +T++ LSS+ L+G I + L L++L+LS+N L G LP L L L+
Sbjct: 521 NLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSHNILKGILPSELSNCHKLSELDASH 580
Query: 421 NNLTGLVPSELLERSKTGSLSL 442
N L G +PS L ++ LSL
Sbjct: 581 NLLNGSIPSTLGSLTELTKLSL 602
Score = 50.4 bits (119), Expect = 2e-05
Identities = 28/67 (41%), Positives = 43/67 (63%), Gaps = 1/67 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ SLNLSS+ L G++ + KL ML+ LD+S+N+L+G L L ++SL +N+ N +
Sbjct: 644 LRSLNLSSNKLNGQLPIDLGKLKMLEELDVSHNNLSGTL-RVLSTIQSLTFINISHNLFS 702
Query: 425 GLVPSEL 431
G VP L
Sbjct: 703 GPVPPSL 709
Score = 49.7 bits (117), Expect = 3e-05
Identities = 28/75 (37%), Positives = 45/75 (59%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L+L ++ L+GE+ SI K+ LQ L L N+L+G LP + +L+ L L + +N+ TG++
Sbjct: 361 LHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYENHFTGVI 420
Query: 428 PSELLERSKTGSLSL 442
P +L S L L
Sbjct: 421 PQDLGANSSLEVLDL 435
Score = 47.8 bits (112), Expect = 1e-04
Identities = 24/69 (34%), Positives = 39/69 (55%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P + ++ + +GL G I S+I ++ L L L +N +GP+P L + +LQ L + NN
Sbjct: 164 PHLETVYFTGNGLNGSIPSNIGNMSELTTLWLDDNQFSGPVPSSLGNITTLQELYLNDNN 223
Query: 423 LTGLVPSEL 431
L G +P L
Sbjct: 224 LVGTLPVTL 232
Score = 46.2 bits (108), Expect = 4e-04
Identities = 26/75 (34%), Positives = 41/75 (54%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
++LSS+ TG I ++ L L+ L L NSL GP P+ L+ + L+ + N L G +
Sbjct: 121 IDLSSNSFTGNIPDTLGALQNLRNLSLFFNSLIGPFPESLLSIPHLETVYFTGNGLNGSI 180
Query: 428 PSELLERSKTGSLSL 442
PS + S+ +L L
Sbjct: 181 PSNIGNMSELTTLWL 195
Score = 45.8 bits (107), Expect = 5e-04
Identities = 27/72 (37%), Positives = 39/72 (53%)
Query: 369 NLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVP 428
+LS + TG I S+ L + + LS+N L+G +P L L L+ LN+ N L G++P
Sbjct: 505 DLSGNNFTGPIPPSLGNLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSHNILKGILP 564
Query: 429 SELLERSKTGSL 440
SEL K L
Sbjct: 565 SELSNCHKLSEL 576
Score = 45.1 bits (105), Expect = 8e-04
Identities = 29/90 (32%), Positives = 46/90 (50%), Gaps = 4/90 (4%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L L + L G + + K +L + DLS N+ GP+P L L+++ + + N L+G +
Sbjct: 481 LILEENNLRGGLPDFVEKQNLL-FFDLSGNNFTGPIPPSLGNLKNVTAIYLSSNQLSGSI 539
Query: 428 PSELLERSKTGSLSLR**SRPL*KGIMQKE 457
P EL K L+L S + KGI+ E
Sbjct: 540 PPELGSLVKLEHLNL---SHNILKGILPSE 566
Score = 42.7 bits (99), Expect = 0.004
Identities = 25/62 (40%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ +L L + L G+I + L L+ L+LS+N LNG LP L +L+ L+ L+V NNL
Sbjct: 620 KLLNLQLGGNLLAGDIPP-VGALQALRSLNLSSNKLNGQLPIDLGKLKMLEELDVSHNNL 678
Query: 424 TG 425
+G
Sbjct: 679 SG 680
Score = 42.4 bits (98), Expect = 0.005
Identities = 21/71 (29%), Positives = 37/71 (51%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + L L+ + L G + +++ L L YLD+ NNSL G +P + + + +++
Sbjct: 210 NITTLQELYLNDNNLVGTLPVTLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSN 269
Query: 421 NNLTGLVPSEL 431
N TG +P L
Sbjct: 270 NQFTGGLPPGL 280
Score = 42.0 bits (97), Expect = 0.007
Identities = 24/68 (35%), Positives = 36/68 (52%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ SL L + TG I + + L+ LDL+ N G +P L + L+ L +G N L
Sbjct: 405 QLVSLALYENHFTGVIPQDLGANSSLEVLDLTRNMFTGHIPPNLCSQKKLKRLLLGYNYL 464
Query: 424 TGLVPSEL 431
G VPS+L
Sbjct: 465 EGSVPSDL 472
Score = 41.6 bits (96), Expect = 0.009
Identities = 25/79 (31%), Positives = 39/79 (48%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ +L L+ + +G I + K + L L N L G +P L L LQ L++ NNL
Sbjct: 309 KLDTLYLAGNHFSGRIPPELGKCKSMIDLQLQQNQLEGEIPGELGMLSQLQYLHLYTNNL 368
Query: 424 TGLVPSELLERSKTGSLSL 442
+G VP + + SL L
Sbjct: 369 SGEVPLSIWKIQSLQSLQL 387
Score = 40.0 bits (92), Expect = 0.026
Identities = 20/68 (29%), Positives = 37/68 (54%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
+I +++LS++ TG + + T L+ + +L+GP+P QL L L + N+
Sbjct: 261 QIDTISLSNNQFTGGLPPGLGNCTSLREFGAFSCALSGPIPSCFGQLTKLDTLYLAGNHF 320
Query: 424 TGLVPSEL 431
+G +P EL
Sbjct: 321 SGRIPPEL 328
Score = 39.7 bits (91), Expect = 0.034
Identities = 22/68 (32%), Positives = 37/68 (54%), Gaps = 1/68 (1%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L L + L G + S + + L+ L L N+L G LPDF ++ ++L ++ NN
Sbjct: 453 KLKRLLLGYNYLEGSVPSDLGGCSTLERLILEENNLRGGLPDF-VEKQNLLFFDLSGNNF 511
Query: 424 TGLVPSEL 431
TG +P L
Sbjct: 512 TGPIPPSL 519
Score = 34.7 bits (78), Expect = 1.1
Identities = 19/67 (28%), Positives = 33/67 (48%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +L+L + L G S+ + L+ + + N LNG +P + + L L + N +
Sbjct: 142 LRNLSLFFNSLIGPFPESLLSIPHLETVYFTGNGLNGSIPSNIGNMSELTTLWLDDNQFS 201
Query: 425 GLVPSEL 431
G VPS L
Sbjct: 202 GPVPSSL 208
Score = 33.9 bits (76), Expect = 1.8
Identities = 23/83 (27%), Positives = 36/83 (42%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
NN + L++ ++ L G I + + LSNN G LP L SL+
Sbjct: 233 NNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSNNQFTGGLPPGLGNCTSLREFGAF 292
Query: 420 KNNLTGLVPSELLERSKTGSLSL 442
L+G +PS + +K +L L
Sbjct: 293 SCALSGPIPSCFGQLTKLDTLYL 315
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 126 bits (317), Expect = 2e-28
Identities = 67/207 (32%), Positives = 120/207 (57%), Gaps = 5/207 (2%)
Query: 621 HAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFG 680
H ++ W+ R+ I + AA GL +LH+ + ++RD K SNILLD N AK++DFG
Sbjct: 180 HLFRRNDPFPWDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSDFG 238
Query: 681 LSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKA--LV 738
L++ D SH++TR GT+GY P++ TG+ K+D+++FG+VLLE++TG A
Sbjct: 239 LAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTK 298
Query: 739 RASGESIHILQWVTP-IVERGDIRSIIDARLQGKFDINSAWKVVEIAMSSTSPIEVERPD 797
R G+ ++ W+ P + + ++ I+D ++G++ A ++ I +S P RP
Sbjct: 299 RPRGQE-SLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPH 357
Query: 798 MSQILAELKECLSLDMVHRNNGRERAI 824
M +++ L+ L++V + ++A+
Sbjct: 358 MKEVVEVLEHIQGLNVVPNRSSTKQAV 384
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 123 bits (309), Expect = 2e-27
Identities = 70/183 (38%), Positives = 112/183 (60%), Gaps = 7/183 (3%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
LNW R IA+ AA GL +LH+ C P +HRD+K SN+LLDEN+ A+++DFG++R +
Sbjct: 983 LNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLM-SA 1041
Query: 689 IDSHISTRP-AGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHI 747
+D+H+S AGT GYV P++ ++ + K D+YS+G+VLLEL+TGK+ A ++
Sbjct: 1042 MDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNL 1101
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVE---IAMSSTSPIEVERPDMSQILAE 804
+ WV + +G I + D L K D + ++++ +A + +RP M Q++A
Sbjct: 1102 VGWV-KLHAKGKITDVFDRELL-KEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
Query: 805 LKE 807
KE
Sbjct: 1160 FKE 1162
Score = 57.4 bits (137), Expect = 2e-07
Identities = 37/88 (42%), Positives = 49/88 (55%), Gaps = 2/88 (2%)
Query: 357 DDDNNPPRITSLNLSSSGLTGEISSSISKLTM--LQYLDLSNNSLNGPLPDFLIQLRSLQ 414
D +N P++ +L++SS+ LTG I S I K M L+ L L NN GP+PD L L
Sbjct: 395 DSFSNLPKLETLDMSSNNLTGIIPSGICKDPMNNLKVLYLQNNLFKGPIPDSLSNCSQLV 454
Query: 415 VLNVGKNNLTGLVPSELLERSKTGSLSL 442
L++ N LTG +PS L SK L L
Sbjct: 455 SLDLSFNYLTGSIPSSLGSLSKLKDLIL 482
Score = 56.2 bits (134), Expect = 3e-07
Identities = 27/67 (40%), Positives = 45/67 (66%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +L L + LTG I +S+S T L ++ LSNN L+G +P L +L +L +L +G N+++
Sbjct: 501 LENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSIS 560
Query: 425 GLVPSEL 431
G +P+EL
Sbjct: 561 GNIPAEL 567
Score = 53.9 bits (128), Expect = 2e-06
Identities = 33/86 (38%), Positives = 53/86 (61%)
Query: 357 DDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVL 416
D +N ++ SL+LS + LTG I SS+ L+ L+ L L N L+G +P L+ L++L+ L
Sbjct: 445 DSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENL 504
Query: 417 NVGKNNLTGLVPSELLERSKTGSLSL 442
+ N+LTG +P+ L +K +SL
Sbjct: 505 ILDFNDLTGPIPASLSNCTKLNWISL 530
Score = 50.4 bits (119), Expect = 2e-05
Identities = 28/83 (33%), Positives = 52/83 (61%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N ++ ++LS++ L+GEI +S+ +L+ L L L NNS++G +P L +SL L++
Sbjct: 520 SNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLN 579
Query: 420 KNNLTGLVPSELLERSKTGSLSL 442
N L G +P L ++S +++L
Sbjct: 580 TNFLNGSIPPPLFKQSGNIAVAL 602
Score = 46.6 bits (109), Expect = 3e-04
Identities = 23/64 (35%), Positives = 37/64 (56%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ LNL + L+G I + L + LDLS N NG +P+ L L L +++ NNL+
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 425 GLVP 428
G++P
Sbjct: 749 GMIP 752
Score = 43.9 bits (102), Expect = 0.002
Identities = 27/101 (26%), Positives = 51/101 (49%), Gaps = 4/101 (3%)
Query: 344 PVNY--MWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNG 401
P N+ ++ G+ T + N + L+LS + L G I + + L L+L +N L+G
Sbjct: 644 PCNFTRVYRGITQPTFNHNGS--MIFLDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSG 701
Query: 402 PLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
+P L L+++ +L++ N G +P+ L + G + L
Sbjct: 702 MIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDL 742
Score = 42.7 bits (99), Expect = 0.004
Identities = 24/90 (26%), Positives = 42/90 (46%), Gaps = 25/90 (27%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLP-------------------- 404
+ L+LS + +G + S+ + + L+ +D+SNN+ +G LP
Sbjct: 330 VVELDLSYNNFSGMVPESLGECSSLELVDISNNNFSGKLPVDTLLKLSNIKTMVLSFNKF 389
Query: 405 -----DFLIQLRSLQVLNVGKNNLTGLVPS 429
D L L+ L++ NNLTG++PS
Sbjct: 390 VGGLPDSFSNLPKLETLDMSSNNLTGIIPS 419
Score = 40.4 bits (93), Expect = 0.020
Identities = 25/79 (31%), Positives = 38/79 (47%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L L + L+GEI + L L+ L L N L GP+P L L +++ N L
Sbjct: 476 KLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQL 535
Query: 424 TGLVPSELLERSKTGSLSL 442
+G +P+ L S L L
Sbjct: 536 SGEIPASLGRLSNLAILKL 554
Score = 38.1 bits (87), Expect = 0.098
Identities = 17/41 (41%), Positives = 29/41 (70%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPD 405
+ L+LS + G I +S++ LT+L +DLSNN+L+G +P+
Sbjct: 713 VAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLSGMIPE 753
Score = 35.4 bits (80), Expect = 0.63
Identities = 15/41 (36%), Positives = 28/41 (67%)
Query: 392 LDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELL 432
LDLS N+ +G +P+ L + SL+++++ NN +G +P + L
Sbjct: 333 LDLSYNNFSGMVPESLGECSSLELVDISNNNFSGKLPVDTL 373
Score = 35.4 bits (80), Expect = 0.63
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 1/64 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ L+LS++ + + S + LQ+LDLS+N G + L L LN+ N
Sbjct: 236 LSYLDLSANNFS-TVFPSFKDCSNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQFV 294
Query: 425 GLVP 428
GLVP
Sbjct: 295 GLVP 298
Score = 35.4 bits (80), Expect = 0.63
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 3/73 (4%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQL-RSLQVLNVGKNN 422
+++ LNL+++ G + S+ LQYL L N G P+ L L +++ L++ NN
Sbjct: 282 KLSFLNLTNNQFVGLVPKLPSE--SLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNN 339
Query: 423 LTGLVPSELLERS 435
+G+VP L E S
Sbjct: 340 FSGMVPESLGECS 352
Score = 32.7 bits (73), Expect = 4.1
Identities = 23/64 (35%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 365 ITSLNLSSSGLTGEIS--SSISKLTMLQYLDLSNNSLNGPLPDFLIQLR-SLQVLNVGKN 421
+ S++L+ + ++G IS SS + L+ L+LS N L+ P + L SLQVL++ N
Sbjct: 136 LDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLKGATFSLQVLDLSYN 195
Query: 422 NLTG 425
N++G
Sbjct: 196 NISG 199
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 123 bits (309), Expect = 2e-27
Identities = 70/183 (38%), Positives = 112/183 (60%), Gaps = 7/183 (3%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
LNW R IA+ AA GL +LH+ C P +HRD+K SN+LLDEN+ A+++DFG++R +
Sbjct: 983 LNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLM-SA 1041
Query: 689 IDSHISTRP-AGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHI 747
+D+H+S AGT GYV P++ ++ + K D+YS+G+VLLEL+TGK+ A ++
Sbjct: 1042 MDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNL 1101
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVE---IAMSSTSPIEVERPDMSQILAE 804
+ WV + +G I + D L K D + ++++ +A + +RP M Q++A
Sbjct: 1102 VGWV-KLHAKGKITDVFDRELL-KEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
Query: 805 LKE 807
KE
Sbjct: 1160 FKE 1162
Score = 56.2 bits (134), Expect = 3e-07
Identities = 27/67 (40%), Positives = 45/67 (66%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +L L + LTG I +S+S T L ++ LSNN L+G +P L +L +L +L +G N+++
Sbjct: 501 LENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSIS 560
Query: 425 GLVPSEL 431
G +P+EL
Sbjct: 561 GNIPAEL 567
Score = 53.9 bits (128), Expect = 2e-06
Identities = 33/86 (38%), Positives = 53/86 (61%)
Query: 357 DDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVL 416
D +N ++ SL+LS + LTG I SS+ L+ L+ L L N L+G +P L+ L++L+ L
Sbjct: 445 DSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENL 504
Query: 417 NVGKNNLTGLVPSELLERSKTGSLSL 442
+ N+LTG +P+ L +K +SL
Sbjct: 505 ILDFNDLTGPIPASLSNCTKLNWISL 530
Score = 53.9 bits (128), Expect = 2e-06
Identities = 36/88 (40%), Positives = 48/88 (53%), Gaps = 2/88 (2%)
Query: 357 DDDNNPPRITSLNLSSSGLTGEISSSISKLTM--LQYLDLSNNSLNGPLPDFLIQLRSLQ 414
D +N ++ +L++SS+ LTG I S I K M L+ L L NN GP+PD L L
Sbjct: 395 DSFSNLLKLETLDMSSNNLTGVIPSGICKDPMNNLKVLYLQNNLFKGPIPDSLSNCSQLV 454
Query: 415 VLNVGKNNLTGLVPSELLERSKTGSLSL 442
L++ N LTG +PS L SK L L
Sbjct: 455 SLDLSFNYLTGSIPSSLGSLSKLKDLIL 482
Score = 50.4 bits (119), Expect = 2e-05
Identities = 28/83 (33%), Positives = 52/83 (61%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N ++ ++LS++ L+GEI +S+ +L+ L L L NNS++G +P L +SL L++
Sbjct: 520 SNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLN 579
Query: 420 KNNLTGLVPSELLERSKTGSLSL 442
N L G +P L ++S +++L
Sbjct: 580 TNFLNGSIPPPLFKQSGNIAVAL 602
Score = 46.6 bits (109), Expect = 3e-04
Identities = 23/64 (35%), Positives = 37/64 (56%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ LNL + L+G I + L + LDLS N NG +P+ L L L +++ NNL+
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 425 GLVP 428
G++P
Sbjct: 749 GMIP 752
Score = 43.9 bits (102), Expect = 0.002
Identities = 27/101 (26%), Positives = 51/101 (49%), Gaps = 4/101 (3%)
Query: 344 PVNY--MWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNG 401
P N+ ++ G+ T + N + L+LS + L G I + + L L+L +N L+G
Sbjct: 644 PCNFTRVYRGITQPTFNHNGS--MIFLDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSG 701
Query: 402 PLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSL 442
+P L L+++ +L++ N G +P+ L + G + L
Sbjct: 702 MIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDL 742
Score = 43.1 bits (100), Expect = 0.003
Identities = 22/63 (34%), Positives = 40/63 (62%), Gaps = 1/63 (1%)
Query: 368 LNLSSSGLTGEIS-SSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGL 426
+++S + +G++ ++SKL+ ++ + LS N G LPD L L+ L++ NNLTG+
Sbjct: 357 VDISYNNFSGKLPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGV 416
Query: 427 VPS 429
+PS
Sbjct: 417 IPS 419
Score = 40.8 bits (94), Expect = 0.015
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQ--LRSLQVLNVGKNN 422
I ++ LS + G + S S L L+ LD+S+N+L G +P + + + +L+VL + N
Sbjct: 379 IKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGVIPSGICKDPMNNLKVLYLQNNL 438
Query: 423 LTGLVPSELLERSKTGSLSL 442
G +P L S+ SL L
Sbjct: 439 FKGPIPDSLSNCSQLVSLDL 458
Score = 40.8 bits (94), Expect = 0.015
Identities = 25/65 (38%), Positives = 35/65 (53%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L L ++ G I S+S + L LDLS N L G +P L L L+ L + N L+G +
Sbjct: 432 LYLQNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEI 491
Query: 428 PSELL 432
P EL+
Sbjct: 492 PQELM 496
Score = 40.4 bits (93), Expect = 0.020
Identities = 25/79 (31%), Positives = 38/79 (47%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
++ L L + L+GEI + L L+ L L N L GP+P L L +++ N L
Sbjct: 476 KLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQL 535
Query: 424 TGLVPSELLERSKTGSLSL 442
+G +P+ L S L L
Sbjct: 536 SGEIPASLGRLSNLAILKL 554
Score = 38.1 bits (87), Expect = 0.098
Identities = 17/41 (41%), Positives = 29/41 (70%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPD 405
+ L+LS + G I +S++ LT+L +DLSNN+L+G +P+
Sbjct: 713 VAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLSGMIPE 753
Score = 35.4 bits (80), Expect = 0.63
Identities = 15/43 (34%), Positives = 29/43 (66%)
Query: 392 LDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLER 434
LDLS N+ +G +P+ L + SL+++++ NN +G +P + L +
Sbjct: 333 LDLSYNNFSGMVPESLGECSSLELVDISYNNFSGKLPVDTLSK 375
Score = 35.4 bits (80), Expect = 0.63
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 1/64 (1%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
++ L+LS++ + + S + LQ+LDLS+N G + L L LN+ N
Sbjct: 236 LSYLDLSANNFS-TVFPSFKDCSNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQFV 294
Query: 425 GLVP 428
GLVP
Sbjct: 295 GLVP 298
Score = 35.4 bits (80), Expect = 0.63
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 3/73 (4%)
Query: 364 RITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQL-RSLQVLNVGKNN 422
+++ LNL+++ G + S+ LQYL L N G P+ L L +++ L++ NN
Sbjct: 282 KLSFLNLTNNQFVGLVPKLPSE--SLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNN 339
Query: 423 LTGLVPSELLERS 435
+G+VP L E S
Sbjct: 340 FSGMVPESLGECS 352
Score = 33.1 bits (74), Expect = 3.1
Identities = 23/64 (35%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 365 ITSLNLSSSGLTGEIS--SSISKLTMLQYLDLSNNSLNGPLPDFLIQLR-SLQVLNVGKN 421
+ S++L+ + ++G IS SS + L+ L+LS N L+ P + L SLQVL++ N
Sbjct: 136 LDSIDLAENTISGPISDISSFGVCSNLKSLNLSKNFLDPPGKEMLKAATFSLQVLDLSYN 195
Query: 422 NLTG 425
N++G
Sbjct: 196 NISG 199
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 119 bits (297), Expect = 4e-26
Identities = 71/199 (35%), Positives = 104/199 (51%), Gaps = 3/199 (1%)
Query: 610 HFQNFETNAYLHA-VENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILL 668
+ N + +LH V+ L+W RL IA AA GL YLH C+P +HRD+K SNILL
Sbjct: 818 YMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILL 877
Query: 669 DENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLL 728
+ A +ADFGL+R D+H++T GT GY+ P++ + K D+YSFG+VLL
Sbjct: 878 SDTFVAHLADFGLARLI-LPYDTHVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLL 936
Query: 729 ELITGKKAL-VRASGESIHILQWVTPIVERGDIRSIIDARLQGKFDINSAWKVVEIAMSS 787
EL+TG++ + V S ++ WV + I D + K V+EIA
Sbjct: 937 ELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIFDPFIYDKDHAEEMLLVLEIACRC 996
Query: 788 TSPIEVERPDMSQILAELK 806
RP Q+++ L+
Sbjct: 997 LGENPKTRPTTQQLVSWLE 1015
Score = 64.3 bits (155), Expect = 1e-09
Identities = 32/87 (36%), Positives = 54/87 (61%), Gaps = 6/87 (6%)
Query: 349 WEGLNCST------DDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGP 402
W G++C + DD N R+ L L L+G++S S++KL L+ L+L++NSL+G
Sbjct: 66 WVGISCKSSVSLGLDDVNESGRVVELELGRRKLSGKLSESVAKLDQLKVLNLTHNSLSGS 125
Query: 403 LPDFLIQLRSLQVLNVGKNNLTGLVPS 429
+ L+ L +L+VL++ N+ +GL PS
Sbjct: 126 IAASLLNLSNLEVLDLSSNDFSGLFPS 152
Score = 57.0 bits (136), Expect = 2e-07
Identities = 30/83 (36%), Positives = 49/83 (58%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
NN PRI ++L+ + G I I + ++YL L++N+L+G +P L QL +L VL +
Sbjct: 179 NNLPRIREIDLAMNYFDGSIPVGIGNCSSVEYLGLASNNLSGSIPQELFQLSNLSVLALQ 238
Query: 420 KNNLTGLVPSELLERSKTGSLSL 442
N L+G + S+L + S G L +
Sbjct: 239 NNRLSGALSSKLGKLSNLGRLDI 261
Score = 55.8 bits (133), Expect = 5e-07
Identities = 30/64 (46%), Positives = 45/64 (69%), Gaps = 1/64 (1%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
LNL+ + L+G I++S+ L+ L+ LDLS+N +G P LI L SL+VLNV +N+ GL+
Sbjct: 115 LNLTHNSLSGSIAASLLNLSNLEVLDLSSNDFSGLFPS-LINLPSLRVLNVYENSFHGLI 173
Query: 428 PSEL 431
P+ L
Sbjct: 174 PASL 177
Score = 54.3 bits (129), Expect = 1e-06
Identities = 25/62 (40%), Positives = 43/62 (69%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
LNL ++ L+G I +++S +T L+ LDLS+N+L+G +P L++L L +V N L+G +
Sbjct: 562 LNLKNNNLSGNIPANLSGMTSLEVLDLSHNNLSGNIPPSLVKLSFLSTFSVAYNKLSGPI 621
Query: 428 PS 429
P+
Sbjct: 622 PT 623
Score = 50.8 bits (120), Expect = 1e-05
Identities = 30/81 (37%), Positives = 46/81 (56%), Gaps = 3/81 (3%)
Query: 362 PPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKN 421
PP I +LS + L G I L L L+L NN+L+G +P L + SL+VL++ N
Sbjct: 535 PPMI---DLSYNSLNGSIWPEFGDLRQLHVLNLKNNNLSGNIPANLSGMTSLEVLDLSHN 591
Query: 422 NLTGLVPSELLERSKTGSLSL 442
NL+G +P L++ S + S+
Sbjct: 592 NLSGNIPPSLVKLSFLSTFSV 612
Score = 49.3 bits (116), Expect = 4e-05
Identities = 32/108 (29%), Positives = 52/108 (47%), Gaps = 8/108 (7%)
Query: 342 CAPVNYMWEGLNCSTDDDNNPPRITSLN------LSSSGLTGEISSSISKLTMLQYLDLS 395
C+ V Y+ GL + + P + L+ L ++ L+G +SS + KL+ L LD+S
Sbjct: 205 CSSVEYL--GLASNNLSGSIPQELFQLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDIS 262
Query: 396 NNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSELLERSKTGSLSLR 443
+N +G +PD ++L L + N G +P L LSLR
Sbjct: 263 SNKFSGKIPDVFLELNKLWYFSAQSNLFNGEMPRSLSNSRSISLLSLR 310
Score = 45.8 bits (107), Expect = 5e-04
Identities = 26/76 (34%), Positives = 41/76 (53%)
Query: 361 NPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGK 420
N + L L+S+ L+G I + +L+ L L L NN L+G L L +L +L L++
Sbjct: 204 NCSSVEYLGLASNNLSGSIPQELFQLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDISS 263
Query: 421 NNLTGLVPSELLERSK 436
N +G +P LE +K
Sbjct: 264 NKFSGKIPDVFLELNK 279
Score = 45.1 bits (105), Expect = 8e-04
Identities = 36/128 (28%), Positives = 58/128 (45%), Gaps = 10/128 (7%)
Query: 309 KDFSQLETQQDDVDNITNIKNAYGVTRNWQGDPCAPVNYMWEGLNCSTDDDNNPPRITSL 368
K+F L + +I NI +A + ++ C + + LN ++ + P +
Sbjct: 371 KNFQSLTSLSFSNSSIQNISSALEILQH-----CQNLKTLVLTLNFQKEELPSVPSLQFK 425
Query: 369 NL-----SSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNL 423
NL +S L G + +S LQ LDLS N L+G +P +L L SL L++ N
Sbjct: 426 NLKVLIIASCQLRGTVPQWLSNSPSLQLLDLSWNQLSGTIPPWLGSLNSLFYLDLSNNTF 485
Query: 424 TGLVPSEL 431
G +P L
Sbjct: 486 IGEIPHSL 493
Score = 44.3 bits (103), Expect = 0.001
Identities = 25/84 (29%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Query: 361 NPPRITSLNLSSSGLTGEISSSI-SKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
N P + LN+ + G I +S+ + L ++ +DL+ N +G +P + S++ L +
Sbjct: 155 NLPSLRVLNVYENSFHGLIPASLCNNLPRIREIDLAMNYFDGSIPVGIGNCSSVEYLGLA 214
Query: 420 KNNLTGLVPSELLERSKTGSLSLR 443
NNL+G +P EL + S L+L+
Sbjct: 215 SNNLSGSIPQELFQLSNLSVLALQ 238
Score = 42.0 bits (97), Expect = 0.007
Identities = 23/54 (42%), Positives = 32/54 (58%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSL 413
+N P + L+LS + L+G I + L L YLDLSNN+ G +P L L+SL
Sbjct: 446 SNSPSLQLLDLSWNQLSGTIPPWLGSLNSLFYLDLSNNTFIGEIPHSLTSLQSL 499
Score = 41.6 bits (96), Expect = 0.009
Identities = 25/82 (30%), Positives = 41/82 (49%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N I+ L+L ++ L+G+I + S +T L LDL++NS +G +P L L+ +N
Sbjct: 299 SNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSLDLASNSFSGSIPSNLPNCLRLKTINFA 358
Query: 420 KNNLTGLVPSELLERSKTGSLS 441
K +P SLS
Sbjct: 359 KIKFIAQIPESFKNFQSLTSLS 380
Score = 38.5 bits (88), Expect = 0.075
Identities = 19/60 (31%), Positives = 33/60 (54%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L++SS+ +G+I +L L Y +N NG +P L RS+ +L++ N L+G +
Sbjct: 259 LDISSNKFSGKIPDVFLELNKLWYFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQI 318
Score = 33.5 bits (75), Expect = 2.4
Identities = 18/60 (30%), Positives = 32/60 (53%)
Query: 372 SSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLVPSEL 431
S+ GE+ S+S + L L NN+L+G + + +L L++ N+ +G +PS L
Sbjct: 287 SNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSLDLASNSFSGSIPSNL 346
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 117 bits (294), Expect = 1e-25
Identities = 66/202 (32%), Positives = 108/202 (52%), Gaps = 10/202 (4%)
Query: 610 HFQNFETNAYLHAVENSNMLNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLD 669
+ +N ++YL + LNWNER +I A GL YLH + +HRDLK SNILLD
Sbjct: 603 YLENLSLDSYLFGKTRRSKLNWNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLD 662
Query: 670 ENMHAKIADFGLSRAFDNDIDSHISTRPAGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLE 729
+NM KI+DFG++R F+ D + + GT+GY+ P++ G ++K+D++SFG+++LE
Sbjct: 663 KNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLE 722
Query: 730 LITGKK--ALVRASGESIHILQWVTPIVERGDIRSIIDARLQGK-------FDINSAWKV 780
+++GKK E+ +L +V + G I+D + F K
Sbjct: 723 IVSGKKNRGFYNLDYEN-DLLSYVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKC 781
Query: 781 VEIAMSSTSPIEVERPDMSQIL 802
++I + + RP MS ++
Sbjct: 782 IQIGLLCVQELAEHRPAMSSVV 803
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 117 bits (294), Expect = 1e-25
Identities = 66/185 (35%), Positives = 110/185 (58%), Gaps = 11/185 (5%)
Query: 629 LNWNERLNIAVDAAHGLDYLHNGCKPPTMHRDLKPSNILLDENMHAKIADFGLSRAFDND 688
LNW+ R IA+ +A GL +LH+ C P +HRD+K SN+LLDEN+ A+++DFG++R +
Sbjct: 978 LNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLM-SA 1036
Query: 689 IDSHISTRP-AGTFGYVDPKFQRTGNTNKKNDIYSFGIVLLELITGKKALVRASGESIHI 747
+D+H+S AGT GYV P++ ++ + K D+YS+G+VLLEL+TGK+ ++
Sbjct: 1037 MDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDNNL 1096
Query: 748 LQWVTPIVERGDIRSIIDARLQGKFDINSAWKV-----VEIAMSSTSPIEVERPDMSQIL 802
+ WV + I + D L + + A ++ +++A++ RP M Q++
Sbjct: 1097 VGWVKQHAKL-RISDVFDPELMKE---DPALEIELLQHLKVAVACLDDRAWRRPTMVQVM 1152
Query: 803 AELKE 807
A KE
Sbjct: 1153 AMFKE 1157
Score = 55.1 bits (131), Expect = 8e-07
Identities = 26/67 (38%), Positives = 43/67 (63%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ +L L + LTGEI S +S T L ++ LSNN L G +P ++ +L +L +L + N+ +
Sbjct: 490 LETLILDFNDLTGEIPSGLSNCTNLNWISLSNNRLTGEIPKWIGRLENLAILKLSNNSFS 549
Query: 425 GLVPSEL 431
G +P+EL
Sbjct: 550 GNIPAEL 556
Score = 49.7 bits (117), Expect = 3e-05
Identities = 30/83 (36%), Positives = 49/83 (58%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N + SL+LS + L+G I SS+ L+ L+ L L N L G +P L+ +++L+ L +
Sbjct: 437 SNCSELVSLHLSFNYLSGTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILD 496
Query: 420 KNNLTGLVPSELLERSKTGSLSL 442
N+LTG +PS L + +SL
Sbjct: 497 FNDLTGEIPSGLSNCTNLNWISL 519
Score = 48.9 bits (115), Expect = 6e-05
Identities = 28/76 (36%), Positives = 44/76 (57%)
Query: 360 NNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVG 419
+N + ++LS++ LTGEI I +L L L LSNNS +G +P L RSL L++
Sbjct: 509 SNCTNLNWISLSNNRLTGEIPKWIGRLENLAILKLSNNSFSGNIPAELGDCRSLIWLDLN 568
Query: 420 KNNLTGLVPSELLERS 435
N G +P+ + ++S
Sbjct: 569 TNLFNGTIPAAMFKQS 584
Score = 48.5 bits (114), Expect = 7e-05
Identities = 27/92 (29%), Positives = 49/92 (52%), Gaps = 2/92 (2%)
Query: 340 DPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSL 399
+PC + ++ G T D+N + L++S + L+G I I + L L+L +N +
Sbjct: 633 NPCNITSRVYGGHTSPTFDNNGS--MMFLDMSYNMLSGYIPKEIGSMPYLFILNLGHNDI 690
Query: 400 NGPLPDFLIQLRSLQVLNVGKNNLTGLVPSEL 431
+G +PD + LR L +L++ N L G +P +
Sbjct: 691 SGSIPDEVGDLRGLNILDLSSNKLDGRIPQAM 722
Score = 48.5 bits (114), Expect = 7e-05
Identities = 25/64 (39%), Positives = 42/64 (65%), Gaps = 2/64 (3%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L++S + L+G+ S +IS T L+ L++S+N GP+P + L+SLQ L++ +N TG +
Sbjct: 250 LDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPP--LPLKSLQYLSLAENKFTGEI 307
Query: 428 PSEL 431
P L
Sbjct: 308 PDFL 311
Score = 47.0 bits (110), Expect = 2e-04
Identities = 23/66 (34%), Positives = 37/66 (55%)
Query: 363 PRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
P + LNL + ++G I + L L LDLS+N L+G +P + L L +++ NN
Sbjct: 678 PYLFILNLGHNDISGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNN 737
Query: 423 LTGLVP 428
L+G +P
Sbjct: 738 LSGPIP 743
Score = 45.4 bits (106), Expect = 6e-04
Identities = 21/38 (55%), Positives = 30/38 (78%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPD 405
L+LSS+ L G I ++S LTML +DLSNN+L+GP+P+
Sbjct: 707 LDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPIPE 744
Score = 43.1 bits (100), Expect = 0.003
Identities = 25/68 (36%), Positives = 38/68 (55%)
Query: 365 ITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLT 424
+ L L ++G TG+I ++S + L L LS N L+G +P L L L+ L + N L
Sbjct: 418 LQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNYLSGTIPSSLGSLSKLRDLKLWLNMLE 477
Query: 425 GLVPSELL 432
G +P EL+
Sbjct: 478 GEIPQELM 485
Score = 42.0 bits (97), Expect = 0.007
Identities = 27/74 (36%), Positives = 42/74 (56%), Gaps = 2/74 (2%)
Query: 365 ITSLNLSSSGLTGEIS-SSISKLTMLQYLDLSNNSLNGPLPDFLIQL-RSLQVLNVGKNN 422
+ SL LSS+ +GE+ ++ K+ L+ LDLS N +G LP+ L L SL L++ NN
Sbjct: 342 LESLALSSNNFSGELPMDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNN 401
Query: 423 LTGLVPSELLERSK 436
+G + L + K
Sbjct: 402 FSGPILPNLCQNPK 415
Score = 42.0 bits (97), Expect = 0.007
Identities = 31/91 (34%), Positives = 50/91 (54%), Gaps = 15/91 (16%)
Query: 340 DPCAPVNYMWEGLNCSTDDDNNPPRITSLNLSSSGLT---GEISSSISKLTMLQYLDLSN 396
+PC ++G+ C D ++TS++LSS L +SSS+ LT L+ L LSN
Sbjct: 60 NPCT-----FDGVTCRDD------KVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSN 108
Query: 397 NSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
+ +NG + F SL L++ +N+L+G V
Sbjct: 109 SHINGSVSGFKCS-ASLTSLDLSRNSLSGPV 138
Score = 40.0 bits (92), Expect = 0.026
Identities = 27/80 (33%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Query: 365 ITSLNLSSSGLTGEISSSISK--LTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNN 422
+ +L+LSS+ +G I ++ + LQ L L NN G +P L L L++ N
Sbjct: 392 LLTLDLSSNNFSGPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNY 451
Query: 423 LTGLVPSELLERSKTGSLSL 442
L+G +PS L SK L L
Sbjct: 452 LSGTIPSSLGSLSKLRDLKL 471
Score = 37.7 bits (86), Expect = 0.13
Identities = 25/84 (29%), Positives = 50/84 (58%), Gaps = 10/84 (11%)
Query: 345 VNYMWEGLNCSTDDDNNPPRITSLNLSSSGLTGEIS--SSISKLTMLQYLDLSNNSLNGP 402
+N G CS +TSL+LS + L+G ++ +S+ + L++L++S+N+L+ P
Sbjct: 111 INGSVSGFKCSAS-------LTSLDLSRNSLSGPVTTLTSLGSCSGLKFLNVSSNTLDFP 163
Query: 403 -LPDFLIQLRSLQVLNVGKNNLTG 425
++L SL+VL++ N+++G
Sbjct: 164 GKVSGGLKLNSLEVLDLSANSISG 187
Score = 36.2 bits (82), Expect = 0.37
Identities = 23/66 (34%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 368 LNLSSSGLTGEISSSIS-KLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGL 426
L+L+ + TGEI +S L LDLS N G +P F L+ L + NN +G
Sbjct: 296 LSLAENKFTGEIPDFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALSSNNFSGE 355
Query: 427 VPSELL 432
+P + L
Sbjct: 356 LPMDTL 361
Score = 33.1 bits (74), Expect = 3.1
Identities = 18/61 (29%), Positives = 32/61 (51%), Gaps = 1/61 (1%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTGLV 427
L++SS+ + I + + LQ+LD+S N L+G + L++LN+ N G +
Sbjct: 227 LDVSSNNFSTGIPF-LGDCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPI 285
Query: 428 P 428
P
Sbjct: 286 P 286
Score = 33.1 bits (74), Expect = 3.1
Identities = 18/58 (31%), Positives = 37/58 (63%), Gaps = 3/58 (5%)
Query: 368 LNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGPLPDFLIQLRSLQVLNVGKNNLTG 425
L +S + ++G++ +S+ L++LD+S+N+ + +P FL +LQ L++ N L+G
Sbjct: 205 LAISGNKISGDVD--VSRCVNLEFLDVSSNNFSTGIP-FLGDCSALQHLDISGNKLSG 259
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.343 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,905,263
Number of Sequences: 164201
Number of extensions: 3502722
Number of successful extensions: 19314
Number of sequences better than 10.0: 1711
Number of HSP's better than 10.0 without gapping: 1106
Number of HSP's successfully gapped in prelim test: 607
Number of HSP's that attempted gapping in prelim test: 16045
Number of HSP's gapped (non-prelim): 2564
length of query: 840
length of database: 59,974,054
effective HSP length: 119
effective length of query: 721
effective length of database: 40,434,135
effective search space: 29153011335
effective search space used: 29153011335
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146586.12


