

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146586.11 - phase: 0 /pseudo
(291 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
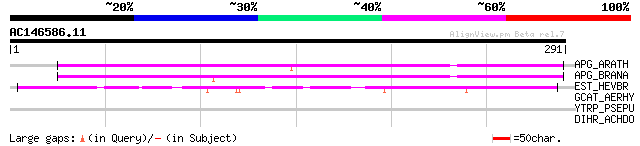
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 179 7e-45
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 174 2e-43
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 110 5e-24
GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase p... 38 0.033
YTRP_PSEPU (P40604) Hypothetical 62.7 kDa protein in trpE-trpG i... 30 5.3
DIHR_ACHDO (Q16983) Diuretic hormone receptor precursor (DH-R) 30 5.3
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 179 bits (454), Expect = 7e-45
Identities = 95/275 (34%), Positives = 149/275 (53%), Gaps = 13/275 (4%)
Query: 26 QDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAE 85
++ +PA+ FGDS D GNN+ L T K+NY PYG DF + TGRF NG +A+D+ A+
Sbjct: 198 ENKTIPAVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAK 257
Query: 86 TLGFTSFAPAYLSPQASGKNLLLGANFASAASGYDEKAATLNHAIPLSQQLEYFKEYQGK 145
+G PAYL P+ +LL G +FAS +GY+ + +AIP+ QL YF++Y K
Sbjct: 258 YMGVKEIVPAYLDPKIQPNDLLTGVSFASGGAGYNPTTSEAANAIPMLDQLTYFQDYIEK 317
Query: 146 L----------AQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSY 195
+ ++AG +K +I + ++ GS+D + Y+ + +D Y++
Sbjct: 318 VNRLVRQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLKNDIDSYTTI 377
Query: 196 LLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKV 255
+ DS +F+ +YG GAR+IGV PPLGC+P+ R + C +N +Q FN K+
Sbjct: 378 IADSAASFVLQLYGYGARRIGVIGTPPLGCVPSQRL---KKKKICNEELNYASQLFNSKL 434
Query: 256 SSAASNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
L K LP V DIY + +++ P+ +
Sbjct: 435 LLILGQLSKTLPNSTFVYMDIYTIISQMLETPAAY 469
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 174 bits (441), Expect = 2e-43
Identities = 94/271 (34%), Positives = 144/271 (52%), Gaps = 9/271 (3%)
Query: 26 QDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDFTNKQPTGRFCNGKLATDFTAE 85
Q+ +PA+ FGDS D GNN+ L T K NY PYG DF TGRF NG++A+D+ ++
Sbjct: 119 QNKTIPAVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYISK 178
Query: 86 TLGFTSFAPAYLSPQASGKN------LLLGANFASAASGYDEKAATLNHAIPLSQQLEYF 139
LG PAY+ + N LL G +FAS +GY + + + QL YF
Sbjct: 179 YLGVKEIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSESWKVTTMLDQLTYF 238
Query: 140 KEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLLDS 199
++Y+ ++ ++ G KK I+ ++ AGS+D + Y+ N + VD +++ + DS
Sbjct: 239 QDYKKRMKKLVGKKKTKKIVSKGAAIVVAGSNDLIYTYFGNGAQHLKNDVDSFTTMMADS 298
Query: 200 FTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGCVARINTDAQGFNKKVSSAA 259
+F+ +YG GAR+IGV PP+GC P+ R + C +N AQ FN K+
Sbjct: 299 AASFVLQLYGYGARRIGVIGTPPIGCTPSQRV---KKKKICNEDLNYAAQLFNSKLVIIL 355
Query: 260 SNLQKQLPGLKIVIFDIYKPLYDLVQNPSNF 290
L K LP IV DIY ++++P ++
Sbjct: 356 GQLSKTLPNSTIVYGDIYSIFSKMLESPEDY 386
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 110 bits (274), Expect = 5e-24
Identities = 93/309 (30%), Positives = 136/309 (43%), Gaps = 48/309 (15%)
Query: 5 NSKETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGRDF 64
N+ + L + C L+ ++ PAI FGDS D G F PPYG F
Sbjct: 7 NNNPIITLSFLLCMLSLAYASETCDFPAIFNFGDSNSDTGGK---AAAFYPLNPPYGETF 63
Query: 65 TNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQAS--GKNLLLGANFASAAS----- 117
++ TGR+ +G+L DF AE SF YLSP S G N GA+FA+A S
Sbjct: 64 FHRS-TGRYSDGRLIIDFIAE-----SFNLPYLSPYLSSLGSNFKHGADFATAGSTIKLP 117
Query: 118 --------GY-----DEKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLY 164
G+ D + + IP SQ + +E G A++ + + +LY
Sbjct: 118 TTIIPAHGGFSPFYLDVQYSQFRQFIPRSQ---FIRETGGIFAELVPEEY---YFEKALY 171
Query: 165 VLSAGSSDFVQNYYTNPWINQAITVDQYSSY---LLDSFTNFIKGVYGLGARKIGVTSLP 221
G +D + + +TV++ ++ L++SF+ +K +Y LGAR + +
Sbjct: 172 TFDIGQNDLTEGFLN-------LTVEEVNATVPDLVNSFSANVKKIYDLGARTFWIHNTG 224
Query: 222 PLGCLPAARTLFGYHEN---GCVARINTDAQGFNKKVSSAASNLQKQLPGLKIVIFDIYK 278
P+GCL T F + E GC N AQ FN K+ + L+K LP V DIY
Sbjct: 225 PIGCLSFILTYFPWAEKDSAGCAKAYNEVAQHFNHKLKEIVAQLRKDLPLATFVHVDIYS 284
Query: 279 PLYDLVQNP 287
Y L P
Sbjct: 285 VKYSLFSEP 293
>GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Glycerophospholipid-cholesterol
acyltransferase) (GCAT)
Length = 335
Score = 37.7 bits (86), Expect = 0.033
Identities = 68/291 (23%), Positives = 112/291 (38%), Gaps = 65/291 (22%)
Query: 7 KETLVLLIVSCFLTCGSFAQDTLVPAIMTFGDSAVDVGN-----NDYLPTLFKANYPPYG 61
K+ V L+ LT + I+ FGDS D G YLP+ PPY
Sbjct: 2 KKWFVCLLGLVALTVQAADSRPAFSRIVMFGDSLSDTGKMYSKMRGYLPSS-----PPYY 56
Query: 62 RDFTNKQPTGRFCNGKLATD-FTAETLGFTSFAPAYLSPQASGKNLLLGANFASAASGYD 120
GRF NG + + T E G T A P A N + ++
Sbjct: 57 E--------GRFSNGPVWLEQLTNEFPGLTIANEAEGGPTAVAYNKI----------SWN 98
Query: 121 EKAATLNHAIPLSQQLEYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTN 180
K +N+ L+Y +V + S D L +L G++D Y
Sbjct: 99 PKYQVINN-------LDY---------EVTQFLQKDSFKPDDLVILWVGAND----YLAY 138
Query: 181 PWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLPAARTLFGYHENGC 240
W +Q + + D+ ++ + GA++I + +LP LG P+AR+
Sbjct: 139 GW-----NTEQDAKRVRDAISDAANRMVLNGAKEILLFNLPDLGQNPSARS--------- 184
Query: 241 VARINTDAQGFNKKVSSAASNLQKQL-PGLKIVIFDIYKPLYDLVQNPSNF 290
++ A + + NL +QL P + +F+I K +++++P NF
Sbjct: 185 -QKVVEAASHVSAYHNQLLLNLARQLAPTGMVKLFEIDKQFAEMLRDPQNF 234
>YTRP_PSEPU (P40604) Hypothetical 62.7 kDa protein in trpE-trpG
intergenic region precursor
Length = 592
Score = 30.4 bits (67), Expect = 5.3
Identities = 60/235 (25%), Positives = 81/235 (33%), Gaps = 44/235 (18%)
Query: 4 MNSKETLVLLIVSCFLTCG-SFAQDTLVPAIMTFGDSAVDVGNNDYLPTLFKANYPPYGR 62
M L + S L C +FA + ++ FGDS D G G
Sbjct: 1 MRKAPLLRFTLASLALACSQAFAAPSPYSGMIVFGDSLSDAGQFG-------------GV 47
Query: 63 DFTNKQPTGRFCNGKLATDFTAETLGFTSFAPAYLSPQASGKNLLLGANFAS--AASGY- 119
FTN G + ++ LG P L P S N +LG + A GY
Sbjct: 48 RFTNLDANGNYA--PVSPMILGGQLGVN---PTELGPSTSPLNPVLGLPDGNNWAVGGYT 102
Query: 120 -----DEKAATLNHAIPLSQQL--EYFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSD 172
D +T IP + + +E G LA + A LY L+ G +D
Sbjct: 103 TQQILDSITSTSETVIPPGRPGAGQVLREKPGYLANGLRADPNA------LYYLTGGGND 156
Query: 173 FVQNYYTNPWINQAITVDQYSSYLLDSFTNFIKGVYGLGARKIGVTSLPPLGCLP 227
F+Q +N + L S +G GAR I V LP LG P
Sbjct: 157 FLQGL-----VNSPADAAAAGARLAASAQALQQG----GARYIMVWLLPDLGQTP 202
>DIHR_ACHDO (Q16983) Diuretic hormone receptor precursor (DH-R)
Length = 441
Score = 30.4 bits (67), Expect = 5.3
Identities = 19/71 (26%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query: 138 YFKEYQGKLAQVAGSKKAASIIKDSLYVLSAGSSDFVQNYYTNPWINQAITVDQYSSYLL 197
YFK+ + + + A I D+ +++SA VQ Y N + + V + YL
Sbjct: 157 YFKDLRCLRNTIHTNLMATYICNDATWIISA----VVQEYVENGGLCSVLAVLMHYFYLT 212
Query: 198 DSFTNFIKGVY 208
+ F F++G+Y
Sbjct: 213 NFFWMFVEGLY 223
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,343,738
Number of Sequences: 164201
Number of extensions: 1451472
Number of successful extensions: 2876
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 2864
Number of HSP's gapped (non-prelim): 6
length of query: 291
length of database: 59,974,054
effective HSP length: 109
effective length of query: 182
effective length of database: 42,076,145
effective search space: 7657858390
effective search space used: 7657858390
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146586.11


