

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.12 - phase: 0 /pseudo
(951 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
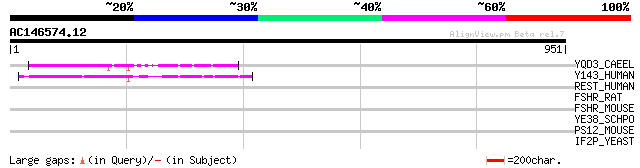
YQD3_CAEEL (Q09263) Hypothetical protein C32D5.3 in chromosome II 81 2e-14
Y143_HUMAN (Q14156) Hypothetical protein KIAA0143 (Fragment) 76 5e-13
REST_HUMAN (P30622) Restin (Cytoplasmic linker protein-170 alpha... 35 1.2
FSHR_RAT (P20395) Follicle stimulating hormone receptor precurso... 33 4.7
FSHR_MOUSE (P35378) Follicle stimulating hormone receptor precur... 33 4.7
YE38_SCHPO (O13892) Hypothetical protein C20G4.08 in chromosome I 32 6.1
PS12_MOUSE (Q80ZC9) Psoriasis susceptibility 1 candidate gene 2 ... 32 6.1
IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B ... 32 8.0
>YQD3_CAEEL (Q09263) Hypothetical protein C32D5.3 in chromosome II
Length = 859
Score = 80.9 bits (198), Expect = 2e-14
Identities = 91/373 (24%), Positives = 161/373 (42%), Gaps = 39/373 (10%)
Query: 32 RYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQRCYKDLRNESFGSV 91
RY++L+ I PR L + KL YA +P ++ +I E L R +DL + V
Sbjct: 13 RYRRLVDSIYPRAVTDGLLYSNMQKLTFYAISHPEKLERIGEYLVMRMVRDLSRQRPVQV 72
Query: 92 KVILCIYRKLLSSCRE--QIPLFASSLLGIIRTLLEQTRADEVRILGCNTLVDFI-IFQT 148
K+ + +LL +C +P F+ + L +++ LLE A ++ L ++ V F I ++
Sbjct: 73 KIAVEAMDQLLQACHSSPSLPQFSENHLRMVQRLLESNNA-KMEQLATDSFVTFSNIEES 131
Query: 149 DGTYMFNLEGFIPKLCQLAQ-----EVGDDERALLLRSAGLQTLSSMV-KFMGEHSHLSM 202
+Y + FI K Q+ GDD R L R AGL+ L +V K + + H ++
Sbjct: 132 SPSYHRQYDFFIDKFSQMCHANPQAAYGDDFR--LARCAGLRGLRGVVWKSVTDDLHPNI 189
Query: 203 ----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATG 258
DKI+ +IL N LQ + K S +Q+ PK + N
Sbjct: 190 WEQQHMDKIVPSILFN---LQEPDDSGK-----GFSSSQI----PKFD-------NTFAD 230
Query: 259 FEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYC 318
++D P S CL + A ++R V+EP+ + D W+ ++
Sbjct: 231 STQSHRVDDEATPKVLSDRCLRELMGKASFG-SLRAVIEPVLKHMDLHKRWTPPP--SFA 287
Query: 319 VLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVA 378
+ ++ + + NS+ ++ L+ HLD + +I I + + +
Sbjct: 288 IHVFRAIIYSIQSQNSYFVIQELINHLD-SMCSADASTRIGIATVLSSIVSIAGTSIGPL 346
Query: 379 VIGAISDLIKHLR 391
++ + L+KHLR
Sbjct: 347 LLSIFNSLLKHLR 359
>Y143_HUMAN (Q14156) Hypothetical protein KIAA0143 (Fragment)
Length = 885
Score = 75.9 bits (185), Expect = 5e-13
Identities = 93/412 (22%), Positives = 170/412 (40%), Gaps = 56/412 (13%)
Query: 15 LCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITEN 74
+C C +LR R YK+L+ I P + L + KL YA P ++ +I
Sbjct: 69 VCCCCSALRPR-------YKRLVDNIFPEDPKDGLVKTDMEKLTFYAVSAPEKLDRIGSY 121
Query: 75 LEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQ-IPLFASSLLGIIRTLLEQTRADEVR 133
L +R +D+ G V + + +LL +C Q I F S L ++ LLE + +++
Sbjct: 122 LAERLSRDVVRHRSGYVLIAMEALDQLLMACHSQSIKPFVESFLHMVAKLLE-SGEPKLQ 180
Query: 134 ILGCNTLVDFIIFQTD-GTYMFNLEGFIPKLCQLAQEV-GDDERALLLRSAGLQTLSSMV 191
+LG N+ V F + D +Y + F+ + + D E +R AG++ + +V
Sbjct: 181 VLGTNSFVKFANIEEDTPSYHRRYDFFVSRFSAMCHSCHSDPEIRTEIRIAGIRGIQGVV 240
Query: 192 -KFMGEHSHLSM----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEE 246
K + + ++ DKI+ ++L N+ K+E+++S+
Sbjct: 241 RKTVNDELRATIWEPQHMDKIVPSLL---------FNMQKIEEVDSRIGPP--------- 282
Query: 247 AHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTE 306
S D +NPA ++ C + A + + P+F + D
Sbjct: 283 -------------SSPSATDKEENPAVLAENCFRELLGRATFG-NMNNAVRPVFAHLDHH 328
Query: 307 NHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQ 366
W + +C + + + A+ SH ++ ++ HLD + P ++ II + +
Sbjct: 329 KLWDPNEFAVHCFKIIMYSIQAQ---YSHHVIQEILGHLDARK-KDAPRVRAGIIQVLLE 384
Query: 367 -VAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDI-GNDAHTLNTKLQS 416
VA K V+ + L+KHLR L EA D+ G ++N S
Sbjct: 385 AVAIAAKGSIGPTVLEVFNTLLKHLR--LSVEFEANDLQGGSVGSVNLNTSS 434
>REST_HUMAN (P30622) Restin (Cytoplasmic linker protein-170 alpha-2)
(CLIP-170) (Reed-Sternberg intermediate filament
associated protein)
Length = 1427
Score = 34.7 bits (78), Expect = 1.2
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 7/93 (7%)
Query: 158 GFIPKLCQLAQEVGDDERALLLRSAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVD 217
G + +L +L ++ D +A +Q + M K E + S++ K +A L+N +D
Sbjct: 1056 GLLQELEELRKQA-DKAKAAQTAEDAMQIMEQMTKEKTE-TLASLEDTKQTNAKLQNELD 1113
Query: 218 LQSKSNLAKVEKLNSQS-----QNQLVQEFPKE 245
++NL VE+LN +NQ ++EF KE
Sbjct: 1114 TLKENNLKNVEELNKSKELLTVENQKMEEFRKE 1146
>FSHR_RAT (P20395) Follicle stimulating hormone receptor precursor
(FSH-R) (Follitropin receptor)
Length = 692
Score = 32.7 bits (73), Expect = 4.7
Identities = 20/70 (28%), Positives = 34/70 (48%)
Query: 436 FDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKAFPDALFHQLLLAMAHP 495
++++ V++ +S I TT+ V T++ +VP L N AF D LL +A
Sbjct: 361 YNILRVLIWFISILAITGNTTVLVVLTTSQYKLTVPRFLMCNLAFADLCIGIYLLLIASV 420
Query: 496 DRETQIGAHS 505
D T+ H+
Sbjct: 421 DIHTKSQYHN 430
>FSHR_MOUSE (P35378) Follicle stimulating hormone receptor precursor
(FSH-R) (Follitropin receptor)
Length = 692
Score = 32.7 bits (73), Expect = 4.7
Identities = 20/70 (28%), Positives = 34/70 (48%)
Query: 436 FDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKAFPDALFHQLLLAMAHP 495
++++ V++ +S I TT+ V T++ +VP L N AF D LL +A
Sbjct: 361 YNILRVLIWFISILAITGNTTVLVVLTTSQYKLTVPRFLMCNLAFADLCIGIYLLLIASV 420
Query: 496 DRETQIGAHS 505
D T+ H+
Sbjct: 421 DIHTKSQYHN 430
>YE38_SCHPO (O13892) Hypothetical protein C20G4.08 in chromosome I
Length = 1076
Score = 32.3 bits (72), Expect = 6.1
Identities = 35/148 (23%), Positives = 66/148 (43%), Gaps = 14/148 (9%)
Query: 123 LLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLRSA 182
LLE A+E ++V + + G LE + K Q+A + +E A +R+A
Sbjct: 751 LLEYKAANEKHTEAILSVVSSTLTENTGKI---LESVVEKSMQVALK---EEIANSVRNA 804
Query: 183 GLQTLSSMVKFMGE-----HSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQ 237
L + F+ + + DFDK S++ + +Q N+A +K + N+
Sbjct: 805 LKNNLEKIESFLENSIAELQNSVREDFDKQTSSLAQLRYSIQ---NVAHAQKESEVKYNE 861
Query: 238 LVQEFPKEEAHVSSMLNVATGFEIESKL 265
L ++ E +V ++L +IE+K+
Sbjct: 862 LNEQVKTLEGYVETVLEKFNDLKIENKV 889
>PS12_MOUSE (Q80ZC9) Psoriasis susceptibility 1 candidate gene 2
protein homolog precursor (SPR1 protein)
Length = 134
Score = 32.3 bits (72), Expect = 6.1
Identities = 14/29 (48%), Positives = 16/29 (54%)
Query: 794 PDDAYPSGPPLFMETPRPGSPLAQIEFPD 822
P D +P PPLF E P PGS + PD
Sbjct: 48 PGDPWPGAPPLFDEPPPPGSNRPWRDLPD 76
>IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B
(eIF-5B) (Translation initiation factor IF-2)
Length = 1002
Score = 32.0 bits (71), Expect = 8.0
Identities = 16/59 (27%), Positives = 35/59 (59%)
Query: 523 KKISKKVESDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVK 581
+++ K + G++++ E SG+ P+ G+ V+E +SL+S ++ L AF +Q+ +
Sbjct: 928 QEVKKGQTAAGVAVRLEDPSGQQPIWGRHVDENDTLYSLVSRRSIDTLKDKAFRDQVAR 986
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 98,331,183
Number of Sequences: 164201
Number of extensions: 3860639
Number of successful extensions: 13629
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 13618
Number of HSP's gapped (non-prelim): 12
length of query: 951
length of database: 59,974,054
effective HSP length: 120
effective length of query: 831
effective length of database: 40,269,934
effective search space: 33464315154
effective search space used: 33464315154
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146574.12


