

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.8 + phase: 0
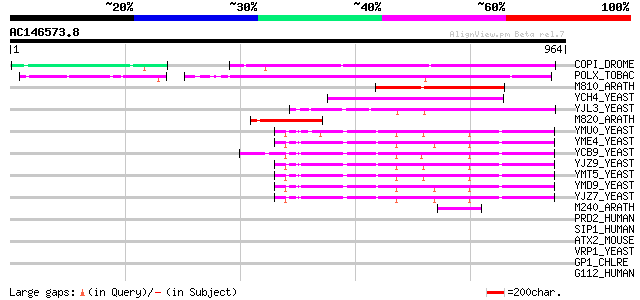
(964 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 367 e-101
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 360 8e-99
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 198 7e-50
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 164 8e-40
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 148 6e-35
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 117 1e-25
YMU0_YEAST (Q04670) Transposon Ty1 protein B 114 2e-24
YME4_YEAST (Q04711) Transposon Ty1 protein B 112 4e-24
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 110 1e-23
YJZ9_YEAST (P47100) Transposon Ty1 protein B 109 3e-23
YMT5_YEAST (Q04214) Transposon Ty1 protein B 108 5e-23
YMD9_YEAST (Q03434) Transposon Ty1 protein B 107 2e-22
YJZ7_YEAST (P47098) Transposon Ty1 protein B 105 6e-22
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 65 9e-10
PRD2_HUMAN (Q13029) PR-domain zinc finger protein 2 (Retinoblast... 41 0.018
SIP1_HUMAN (O60315) Zinc finger homeobox protein 1b (Smad intera... 40 0.023
ATX2_MOUSE (O70305) Ataxin-2 (Spinocerebellar ataxia type 2 prot... 40 0.023
VRP1_YEAST (P37370) Verprolin 39 0.051
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 39 0.051
G112_HUMAN (Q8IZF6) G-protein coupled receptor GPR112 39 0.067
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 367 bits (942), Expect = e-101
Identities = 218/587 (37%), Positives = 332/587 (56%), Gaps = 25/587 (4%)
Query: 382 LSPSTSSGNESSPSPSPTNN---SIALTNPPADSNATNHIITRSQNNIFKPKKFFYASNH 438
L+ S SGN + S T I + NP ++ I RS+ KP+ + ++
Sbjct: 816 LNESKGSGNPNESRESETAEHLKEIGIDNP-TKNDGIEIINRRSERLKTKPQISYNEEDN 874
Query: 439 PLPE---------NLEPSN---VRQAMQHPHWRQAISEEFNALIRNGTWSLVPPPKNKNI 486
L + N P++ ++ W +AI+ E NA N TW++ P+NKNI
Sbjct: 875 SLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRPENKNI 934
Query: 487 VDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTK 546
VD +W+F +K N G YKARLVA+GFTQ +D++ETFAPV R + + IL++ +
Sbjct: 935 VDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQY 994
Query: 547 GWKMHQLDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDAL 606
K+HQ+DV AFL G+L EE+YM P G+ S VCKL+KAIYGL+QA R W +
Sbjct: 995 NLKVHQMDVKTAFLNGTLKEEIYMRLPQGI--SCNSDNVCKLNKAIYGLKQAARCWFEVF 1052
Query: 607 KTFITSHGFTTSQSDPSLFIYASG--TTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRF 664
+ + F S D ++I G Y L+YVDD+++ D + +++F + L +F
Sbjct: 1053 EQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKF 1112
Query: 665 SLKHMGTPHYFLGIELIPSKTVLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTSTLTLND 724
+ + +F+GI + + ++LSQ +++ IL +F+M+ TPL + L +
Sbjct: 1113 RMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYELLN 1172
Query: 725 GTAAADSTLYRQIIGALQYLNL-TRPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKAT 783
+T R +IG L Y+ L TRPDL+ A+N LS++ K S +Q+LKR+LRYLK T
Sbjct: 1173 SDEDC-NTPCRSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGT 1231
Query: 784 INFGILLKKTNALTLQV--YTDADWDGNIDDRTSTSAYLI-YFGGNPISWLSRKQRTVAR 840
I+ ++ KK A ++ Y D+DW G+ DR ST+ YL F N I W +++Q +VA
Sbjct: 1232 IDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAA 1291
Query: 841 SSTEAEYRAVATATAEIMWITNLLSELHIPLSKPPLLLCDNVGATYLCSNPVLHSKMKHI 900
SSTEAEY A+ A E +W+ LL+ ++I L P + DN G + +NP H + KHI
Sbjct: 1292 SSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHI 1351
Query: 901 SLDYHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSKM 947
+ YHF REQV++ + + ++ T++QLAD+ TKPLP A+F ELR K+
Sbjct: 1352 DIKYHFAREQVQNNVICLEYIPTENQLADIFTKPLPAARFVELRDKL 1398
Score = 63.5 bits (153), Expect = 3e-09
Identities = 58/288 (20%), Positives = 110/288 (38%), Gaps = 23/288 (7%)
Query: 3 QFNALLIGYDLFGFVDGTKPEPAEKHVDYNYWRRQDKLILHAILSSVEASIITMLGNVKN 62
+ ALL D+ VDG P + + W++ ++ I+ + S + +
Sbjct: 22 RIRALLAEQDVLKVVDGLMPNEVD-----DSWKKAERCAKSTIIEYLSDSFLNFATSDIT 76
Query: 63 SKDAWDILNKMFASKTRARIMHLKERLTRTSKGSK-SVSEYLQAIKAISDELAIINKPID 121
++ + L+ ++ K+ A + L++RL S+ S+ + + EL I+
Sbjct: 77 ARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELISELLAAGAKIE 136
Query: 122 DDDLVIHAPNGLGSEFKEIAAALRT-RENAIAFDELHDILVDHETFLQRDQEPTIIPTAQ 180
+ D + H L S + I A+ T E + + + L+D E ++ D T
Sbjct: 137 EMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLDQEIKIKNDHNDTSKKVMN 196
Query: 181 VAYRGKPKYHKNGSFHNHINPSVNNISDSQGQQKQVMCQFCDKSGHTAKECY-------- 232
+KN F N + + + +V C C + GH K+C+
Sbjct: 197 AIVHNNNNTYKNNLFKNRVTKPKKIFKGNS--KYKVKCHHCGREGHIKKDCFHYKRILNN 254
Query: 233 ------KIHGYPSKNGVRSAAHSARYTPHTDETDWILDSGATHHLTNN 274
K + +G+ T D ++LDSGA+ HL N+
Sbjct: 255 KNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGASDHLIND 302
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 360 bits (925), Expect = 8e-99
Identities = 219/651 (33%), Positives = 345/651 (52%), Gaps = 36/651 (5%)
Query: 304 DPKTNKIYTSRHVVFHDNHFPYPTINQNAHAANLPINISLIPPHTILSLNPTIPPPQAPN 363
DP K+ SR VVF ++ + A + + +IP TIP
Sbjct: 690 DPVKKKVIRSRDVVFRESEV------RTAADMSEKVKNGIIPNFV------TIPST---- 733
Query: 364 SLMATTPPTPEAILQEAPLSPSTSSGNESSPSPSPTNNSIALTNPPADSNATNHIITRSQ 423
+ P + E+ E +S E + + P + + RS+
Sbjct: 734 ---SNNPTSAESTTDE--VSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEEQHQPLRRSE 788
Query: 424 NNIFKPKKFFYASNHPLPENLEPSNVRQAMQHPHWRQ---AISEEFNALIRNGTWSLVPP 480
+ +++ + ++ EP ++++ + HP Q A+ EE +L +NGT+ LV
Sbjct: 789 RPRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVEL 848
Query: 481 PKNKNIVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLIL 540
PK K + CKW+F++K++ D + YKARLV KGF Q G+DF E F+PVV+ +I+ IL
Sbjct: 849 PKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTIL 908
Query: 541 TIALTKGWKMHQLDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPR 600
++A + ++ QLDV AFL G L EE+YM QP G + + + VCKL+K++YGL+QAPR
Sbjct: 909 SLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPR 968
Query: 601 AWHDALKTFITSHGFTTSQSDPSLFIYA-SGTTLAYFLVYVDDLLLTGNDASFLHHFIQS 659
W+ +F+ S + + SDP ++ S L+YVDD+L+ G D +
Sbjct: 969 QWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGD 1028
Query: 660 LSNRFSLKHMGTPHYFLGIELIPSKT--VLFLSQHKFIRDILQRFDMDAAKPTYTPLSTT 717
LS F +K +G LG++++ +T L+LSQ K+I +L+RF+M AKP TPL+
Sbjct: 1029 LSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGH 1088
Query: 718 STL------TLNDGTAAADSTLYRQIIGALQY-LNLTRPDLSFAINKLSQFMHKPTSLHF 770
L T + Y +G+L Y + TRPD++ A+ +S+F+ P H+
Sbjct: 1089 LKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHW 1148
Query: 771 QHLKRLLRYLKATINFGILLKKTNALTLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISW 830
+ +K +LRYL+ T + ++ + L+ YTDAD G+ID+R S++ YL F G ISW
Sbjct: 1149 EAVKWILRYLRGTTGDCLCFGGSDPI-LKGYTDADMAGDIDNRKSSTGYLFTFSGGAISW 1207
Query: 831 LSRKQRTVARSSTEAEYRAVATATAEIMWITNLLSELHIPLSKPPLLLCDNVGATYLCSN 890
S+ Q+ VA S+TEAEY A E++W+ L EL + K ++ CD+ A L N
Sbjct: 1208 QSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLH-QKEYVVYCDSQSAIDLSKN 1266
Query: 891 PVLHSKMKHISLDYHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFE 941
+ H++ KHI + YH++RE V D L+V +ST + AD+LTK +PR KFE
Sbjct: 1267 SMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVPRNKFE 1317
Score = 57.4 bits (137), Expect = 2e-07
Identities = 52/277 (18%), Positives = 116/277 (41%), Gaps = 31/277 (11%)
Query: 17 VDGTKPEPAEKHVDYNYWRRQDKLILHAILSSVEASIITMLGNVKNSKDAWDILNKMFAS 76
VD KP+ + W D+ AI + ++ + + ++ W L ++ S
Sbjct: 39 VDSKKPDTMKAED----WADLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMS 94
Query: 77 KTRARIMHLKERL--TRTSKGSKSVSEYLQAIKAISDELAIINKPIDDDDLVIHAPNGLG 134
KT ++LK++L S+G+ +S +L + +LA + I+++D I N L
Sbjct: 95 KTLTNKLYLKKQLYALHMSEGTNFLS-HLNVFNGLITQLANLGVKIEEEDKAILLLNSLP 153
Query: 135 SEFKEIAAALRTRENAIAFDELHDILVDHETFLQRDQEPTIIPTAQVAYRGKPKYHKNGS 194
S + +A + + I ++ L+ +E ++ + + G+ + ++ S
Sbjct: 154 SSYDNLATTILHGKTTIELKDVTSALLLNEKMRKKPENQG----QALITEGRGRSYQRSS 209
Query: 195 FHNHINPSVNNISDSQGQQKQVMCQFCDKSGHTAKECYKIH-GYPSKNGVRSAAHSARYT 253
+N+ S ++ + + C C++ GH ++C G +G ++ ++A
Sbjct: 210 -NNYGRSGARGKSKNRSKSRVRNCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMV 268
Query: 254 PHTD------------------ETDWILDSGATHHLT 272
+ D E++W++D+ A+HH T
Sbjct: 269 QNNDNVVLFINEEEECMHLSGPESEWVVDTAASHHAT 305
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 198 bits (503), Expect = 7e-50
Identities = 106/228 (46%), Positives = 144/228 (62%), Gaps = 7/228 (3%)
Query: 635 YFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMGTPHYFLGIELIPSKTVLFLSQHKF 694
Y L+YVDD+LLTG+ + L+ I LS+ FS+K +G HYFLGI++ + LFLSQ K+
Sbjct: 2 YLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKY 61
Query: 695 IRDILQRFDMDAAKPTYTPLSTTSTLTLNDGTAAA---DSTLYRQIIGALQYLNLTRPDL 751
IL M KP TPL L LN + A D + +R I+GALQYL LTRPD+
Sbjct: 62 AEQILNNAGMLDCKPMSTPLP----LKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDI 117
Query: 752 SFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLKKTNALTLQVYTDADWDGNID 811
S+A+N + Q MH+PT F LKR+LRY+K TI G+ + K + L +Q + D+DW G
Sbjct: 118 SYAVNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTS 177
Query: 812 DRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEYRAVATATAEIMW 859
R ST+ + + G N ISW +++Q TV+RSSTE EYRA+A AE+ W
Sbjct: 178 TRRSTTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 164 bits (416), Expect = 8e-40
Identities = 95/308 (30%), Positives = 157/308 (50%), Gaps = 3/308 (0%)
Query: 553 LDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITS 612
+DV+ AFL +++E +Y+ QPPG + + P YV +L+ +YGL+QAP W++ + +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 613 HGFTTSQSDPSLFIYASGTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMGTP 672
GF + + L+ ++ Y VYVDDLL+ Q L+ +S+K +G
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 673 HYFLGIELIPSKT-VLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTSTLTLNDGTAAADS 731
FLG+ + S + LS +I +++ K T TPL + L D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 732 TLYRQIIGALQY-LNLTRPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILL 790
T Y+ I+G L + N RPD+S+ ++ LS+F+ +P ++H + +R+LRYL T + +
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 791 KKTNALTLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQR-TVARSSTEAEYRA 849
+ + + L VY DA D ST Y+ G P++W S+K + + STEAEY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 850 VATATAEI 857
+ EI
Sbjct: 301 ASETVMEI 308
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 148 bits (374), Expect = 6e-35
Identities = 124/482 (25%), Positives = 222/482 (45%), Gaps = 33/482 (6%)
Query: 486 IVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQTIKLILTIALT 545
IV +F KRN YKAR+V +G TQ P + + IK+ L IA
Sbjct: 1322 IVPTNTIFTKKRN-----GIYKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANN 1375
Query: 546 KGWKMHQLDVNNAFLQGSLNEEVYMAQPPGLKDSQQPTYVCKLHKAIYGLRQAPRAWHDA 605
+ M LD+N+AFL L EE+Y+ P + V KL+KA+YGL+Q+P+ W+D
Sbjct: 1376 RNMFMKTLDINHAFLYAKLEEEIYIPHPHDRR------CVVKLNKALYGLKQSPKEWNDH 1429
Query: 606 LKTFITSHGFTTSQSDPSLFIYASGTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFS 665
L+ ++ G + P L Y + VYVDD ++ ++ L FI L + F
Sbjct: 1430 LRQYLNGIGLKDNSYTPGL--YQTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFE 1487
Query: 666 LKHMGT------PHYFLGIELIPSKTV--LFLSQHKFIRDILQRFDMDAAKPTYTPLSTT 717
LK GT LG++L+ +K + + L+ FI + ++++ + K + +
Sbjct: 1488 LKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHM 1547
Query: 718 ST---------LTLNDGTAAADSTLYRQIIGALQYL-NLTRPDLSFAINKLSQFMHKPTS 767
ST L +++ +Q++G L Y+ + R D+ FA+ K+++ ++ P
Sbjct: 1548 STYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHE 1607
Query: 768 LHFQHLKRLLRYLKATINFGILLKKTNALTLQVYTDADWD-GNIDDRTSTSAYLIYFGGN 826
F + ++++YL + GI + +V D G+ D S ++++G N
Sbjct: 1608 RVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDASVGSEYDAQSRIGVILWYGMN 1667
Query: 827 PISWLSRKQRTVARSSTEAEYRAVATATAEIMWITNLLSELHIPLSKPPLLLCDNVGATY 886
+ S K SSTEAE A+ A+ + L EL + +++ D+ A
Sbjct: 1668 IFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQ 1727
Query: 887 LCSNPVLHSKMKHISLDYHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+ K K + ++E++++ +++ ++ K +ADLLTKP+ + F+
Sbjct: 1728 GLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFKRFIQV 1787
Query: 947 MK 948
+K
Sbjct: 1788 LK 1789
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 117 bits (294), Expect = 1e-25
Identities = 64/128 (50%), Positives = 84/128 (65%), Gaps = 5/128 (3%)
Query: 418 IITRSQNNIFK--PKKFFYASNHPLPENLEPSNVRQAMQHPHWRQAISEEFNALIRNGTW 475
++TRS+ I K PK Y+ EP +V A++ P W QA+ EE +AL RN TW
Sbjct: 1 MLTRSKAGINKLNPK---YSLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTW 57
Query: 476 SLVPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLVAKGFTQCPGVDFKETFAPVVRPQT 535
LVPPP N+NI+ CKW+F+ K + DGT+D KARLVAKGF Q G+ F ET++PVVR T
Sbjct: 58 ILVPPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTAT 117
Query: 536 IKLILTIA 543
I+ IL +A
Sbjct: 118 IRTILNVA 125
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 114 bits (284), Expect = 2e-24
Identities = 130/528 (24%), Positives = 232/528 (43%), Gaps = 68/528 (12%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
QA +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 825 QAYHKEVNQLLKMKTWDTDRYYDRKEIDP---KRVINSMFIFNRKR--DGT---HKARFV 876
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIK-----LILTIALTKGWKMHQLDVNNAFLQGSLNE 566
A+G Q P +T+ P ++ T+ L++AL + + QLD+++A+L + E
Sbjct: 877 ARGDIQHP-----DTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKE 931
Query: 567 EVYMAQPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSL 624
E+Y+ PP G+ D + +L K++YGL+Q+ W++ +K+++ S
Sbjct: 932 ELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSC 986
Query: 625 FIYASGTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLG 677
S T+ F VDD++L D + I +L ++ K + + LG
Sbjct: 987 VFKNSQVTICLF---VDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILG 1043
Query: 678 IELIPSK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLST---------TSTLTLNDGTA 727
+E+ + + L + + + + ++ P LS L L +
Sbjct: 1044 LEIKYQRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDY 1102
Query: 728 AADSTLYRQIIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINF 786
+++IG Y+ R DL + IN L+Q + P+ L++++ T +
Sbjct: 1103 KMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDK 1162
Query: 787 GILLKKTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSS 842
++ K+ + L V +DA + GN S + G I S K S+
Sbjct: 1163 QLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTST 1221
Query: 843 TEAEYRAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMK 898
TEAE A+ +E + + N LS L L+K P+ LL D+ + + SN + +
Sbjct: 1222 TEAEIHAI----SESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNR 1277
Query: 899 HISLDYHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1278 FFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 112 bits (281), Expect = 4e-24
Identities = 127/523 (24%), Positives = 226/523 (42%), Gaps = 58/523 (11%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 825 EAYHKEVNQLLKMNTWDTDKYYDRKEIDP---KRVINSMFIFNRKR--DGT---HKARFV 876
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + V + L++AL + + QLD+++A+L + EE+Y+
Sbjct: 877 ARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP G+ D + +L K++YGL+Q+ W++ +K+++ S S
Sbjct: 937 PPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNS 991
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L D + I +L ++ K + + LG+E+
Sbjct: 992 QVTICLF---VDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY 1048
Query: 683 SK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTST--LTLNDGTAAADSTLYRQ--- 736
+ + L + + + + ++ P LS L ++ D Y++
Sbjct: 1049 QRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVH 1107
Query: 737 ----IIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK 791
+IG Y+ R DL + IN L+Q + P+ L++++ T + ++
Sbjct: 1108 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWH 1167
Query: 792 KTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEY 847
K L +DA + GN S + G I S K S+TEAE
Sbjct: 1168 KNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEI 1226
Query: 848 RAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLD 903
A+ +E + + N LS L L+K P+ LL D+ + + SN + +
Sbjct: 1227 HAI----SESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTK 1282
Query: 904 YHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1283 AMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 110 bits (276), Expect = 1e-23
Identities = 135/582 (23%), Positives = 243/582 (41%), Gaps = 62/582 (10%)
Query: 400 NNSIALTNPPADSNATNHIITRSQNNIFKPKKFFYASNHPLPENLEPSNVRQAMQHPHWR 459
N ++ PP N I KP + + + N + + +
Sbjct: 1213 NKNMRSLEPPRSKKRINLIAAIKGVKSIKPVRTTLRYDEAITYN------KDNKEKDRYV 1266
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E + L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 1267 EAYHKEISQLLKMNTWDTNKYYDRNDIDP---KKVINSMFIFNKKR--DGT---HKARFV 1318
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + + V + L+IAL + + QLD+++A+L + EE+Y+
Sbjct: 1319 ARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIR 1378
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP GL D + +L K++YGL+Q+ W++ +K+++ + S S
Sbjct: 1379 PPPHLGLNDK-----LLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVFKNS 1433
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L D + I +L ++ K + + LG+E+
Sbjct: 1434 QVTICLF---VDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY 1490
Query: 683 SKT-VLFLSQHKFIRDILQRFDM---DAAKPTYTP-----LSTTSTLTLNDGTAAADSTL 733
++ + L K + + L + ++ K P L +++
Sbjct: 1491 QRSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELEIDEDEYKEKVHE 1550
Query: 734 YRQIIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLKK 792
+++IG Y+ R DL + IN L+Q + P+ L++++ T + ++ K
Sbjct: 1551 MQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHK 1610
Query: 793 TNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEYR 848
L +DA + GN S + G I S K S+TEAE
Sbjct: 1611 NKPTKPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIH 1669
Query: 849 AVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLDY 904
AV +E + + N LS L L+K P+ LL D+ + + S + +
Sbjct: 1670 AV----SEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKA 1725
Query: 905 HFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1726 MRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 109 bits (273), Expect = 3e-23
Identities = 126/523 (24%), Positives = 226/523 (43%), Gaps = 58/523 (11%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 1252 EAYHKEVNQLLKMKTWDTDEYYDRKEIDP---KRVINSMFIFNKKR--DGT---HKARFV 1303
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + V + L++AL + + QLD+++A+L + EE+Y+
Sbjct: 1304 ARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 1363
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP G+ D + +L K++YGL+Q+ W++ +K+++ S S
Sbjct: 1364 PPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVFKNS 1418
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L + + I+ L ++ K + + LG+E+
Sbjct: 1419 QVTICLF---VDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKY 1475
Query: 683 SK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLST---------TSTLTLNDGTAAADST 732
+ + L + + + + ++ P LS L L +
Sbjct: 1476 QRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVH 1534
Query: 733 LYRQIIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK 791
+++IG Y+ R DL + IN L+Q + P+ L++++ T + ++
Sbjct: 1535 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWH 1594
Query: 792 KTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEY 847
K+ + L V +DA + GN S + G I S K S+TEAE
Sbjct: 1595 KSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEI 1653
Query: 848 RAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLD 903
A+ +E + + N LS L L K P+ LL D+ + + SN + +
Sbjct: 1654 HAI----SESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTK 1709
Query: 904 YHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1710 AMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 108 bits (271), Expect = 5e-23
Identities = 126/523 (24%), Positives = 225/523 (42%), Gaps = 58/523 (11%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 825 EAYHKEVNQLLKMKTWDTDKYYDRKEIDP---KRVINSMFIFNRKR--DGT---HKARFV 876
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + V + L++AL + + QLD+++A+L + EE+Y+
Sbjct: 877 ARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP G+ D + +L K++YGL+Q+ W++ +K+++ S S
Sbjct: 937 PPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFENS 991
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L + + I L ++ K + + LG+E+
Sbjct: 992 QVTICLF---VDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKY 1048
Query: 683 SK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLST---------TSTLTLNDGTAAADST 732
+ + L + + + + ++ P LS L L +
Sbjct: 1049 QRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVH 1107
Query: 733 LYRQIIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK 791
+++IG Y+ R DL + IN L+Q + P+ L++++ T + ++
Sbjct: 1108 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWH 1167
Query: 792 KTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEY 847
K+ + L V +DA + GN S + G I S K S+TEAE
Sbjct: 1168 KSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEI 1226
Query: 848 RAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLD 903
A+ +E + + N LS L L K P+ LL D+ + + SN + +
Sbjct: 1227 HAI----SESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTK 1282
Query: 904 YHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1283 AMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 107 bits (267), Expect = 2e-22
Identities = 126/523 (24%), Positives = 225/523 (42%), Gaps = 58/523 (11%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 825 EAYHKEVNQLLKMKTWDTDEYYDRKEIDP---KRVINSMFIFNKKR--DGT---HKARFV 876
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + V + L++AL + + QLD+++A+L + EE+Y+
Sbjct: 877 ARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP G+ D + +L K++YGL+Q+ W++ +K+++ S S
Sbjct: 937 PPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNS 991
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L D + I +L ++ K + + LG+E+
Sbjct: 992 QVTICLF---VDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY 1048
Query: 683 SK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTST--LTLNDGTAAADSTLYRQ--- 736
+ + L + + + + ++ P LS L ++ D Y++
Sbjct: 1049 QRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVH 1107
Query: 737 ----IIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK 791
+IG Y+ R DL + IN L+Q + P+ L++++ T + ++
Sbjct: 1108 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWH 1167
Query: 792 KTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEY 847
K L +DA + GN S + G I S K S+TEAE
Sbjct: 1168 KNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEI 1226
Query: 848 RAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLD 903
A+ +E + + N LS L L+K P+ LL D+ + + S + +
Sbjct: 1227 HAI----SESVPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTK 1282
Query: 904 YHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1283 AMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 105 bits (262), Expect = 6e-22
Identities = 126/523 (24%), Positives = 224/523 (42%), Gaps = 58/523 (11%)
Query: 460 QAISEEFNALIRNGTWSL--------VPPPKNKNIVDCKWLFRIKRNPDGTIDHYKARLV 511
+A +E N L++ TW + P K +++ ++F KR DGT +KAR V
Sbjct: 1252 EAYHKEVNQLLKMKTWDTDEYYDRKEIDP---KRVINSMFIFNKKR--DGT---HKARFV 1303
Query: 512 AKGFTQCPGVDFKETFAPVVRPQTIKLILTIALTKGWKMHQLDVNNAFLQGSLNEEVYMA 571
A+G Q P + V + L++AL + + QLD+++A+L + EE+Y+
Sbjct: 1304 ARGDIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 1363
Query: 572 QPP--GLKDSQQPTYVCKLHKAIYGLRQAPRAWHDALKTFITSHGFTTSQSDPSLFIYAS 629
PP G+ D + +L K+ YGL+Q+ W++ +K+++ S S
Sbjct: 1364 PPPHLGMNDK-----LIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNS 1418
Query: 630 GTTLAYFLVYVDDLLLTGNDASFLHHFIQSLSNRFSLKHMG-------TPHYFLGIELIP 682
T+ F VDD++L D + I +L ++ K + + LG+E+
Sbjct: 1419 QVTICLF---VDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY 1475
Query: 683 SK-TVLFLSQHKFIRDILQRFDMDAAKPTYTPLSTTST--LTLNDGTAAADSTLYRQ--- 736
+ + L + + + + ++ P LS L ++ D Y++
Sbjct: 1476 QRGKYMKLGMENSLTEKIPKLNV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVH 1534
Query: 737 ----IIGALQYLNLT-RPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLK 791
+IG Y+ R DL + IN L+Q + P+ L++++ T + ++
Sbjct: 1535 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWH 1594
Query: 792 KTNAL----TLQVYTDADWDGNIDDRTSTSAYLIYFGGNPISWLSRKQRTVARSSTEAEY 847
K L +DA + GN S + G I S K S+TEAE
Sbjct: 1595 KNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEI 1653
Query: 848 RAVATATAEIMWITNLLSELHIPLSKPPL---LLCDNVGA-TYLCSNPVLHSKMKHISLD 903
A+ +E + + N LS L L+K P+ LL D+ + + S + +
Sbjct: 1654 HAI----SESVPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTK 1709
Query: 904 YHFVREQVRDGKLQVSHVSTKDQLADLLTKPLPRAKFEELRSK 946
+R++V L V ++ TK +AD++TKPLP F+ L +K
Sbjct: 1710 AMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 65.1 bits (157), Expect = 9e-10
Identities = 29/77 (37%), Positives = 46/77 (59%)
Query: 743 YLNLTRPDLSFAINKLSQFMHKPTSLHFQHLKRLLRYLKATINFGILLKKTNALTLQVYT 802
YL +TRPDL+FA+N+LSQF + Q + ++L Y+K T+ G+ T+ L L+ +
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 803 DADWDGNIDDRTSTSAY 819
D+DW D R S + +
Sbjct: 62 DSDWASCPDTRRSVTGF 78
>PRD2_HUMAN (Q13029) PR-domain zinc finger protein 2 (Retinoblastoma
protein-interacting zinc-finger protein) (MTE-binding
protein) (MTB-ZF) (GATA-3 binding protein G3B)
Length = 1719
Score = 40.8 bits (94), Expect = 0.018
Identities = 31/93 (33%), Positives = 43/93 (45%), Gaps = 10/93 (10%)
Query: 323 FPYPTINQ------NAHAANLPINISLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAI 376
FP PT+ ++ A P S P ++ +P+ PPP P L TPP P +
Sbjct: 934 FPAPTVESTPDVCPSSPALQTPSLSSGQLPPLLIPTDPSSPPP-CPPVLTVATPPPP--L 990
Query: 377 LQEAPLSPSTSSGNESSPSPSPTNNSIALTNPP 409
L PL P+ SS P PSP +N+ A + P
Sbjct: 991 LPTVPL-PAPSSSASPHPCPSPLSNATAQSPLP 1022
Score = 35.0 bits (79), Expect = 0.96
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 7/81 (8%)
Query: 324 PYPTINQNAHAANLPI-NISLIPPHTILSLNPTIPPPQAP----NSLMATTPPTPEAILQ 378
P P+ + + H P+ N + P ILS PT+ P +P LM+ P P +
Sbjct: 997 PAPSSSASPHPCPSPLSNATAQSPLPILS--PTVSPSPSPIPPVEPLMSAASPGPPTLSS 1054
Query: 379 EAPLSPSTSSGNESSPSPSPT 399
+ S S+SS + SS S SP+
Sbjct: 1055 SSSSSSSSSSFSSSSSSSSPS 1075
>SIP1_HUMAN (O60315) Zinc finger homeobox protein 1b (Smad
interacting protein 1) (SMADIP1) (HRIHFB2411)
Length = 1214
Score = 40.4 bits (93), Expect = 0.023
Identities = 31/108 (28%), Positives = 51/108 (46%), Gaps = 13/108 (12%)
Query: 359 PQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPSPSPTNNSIALTNPPADSNATNHI 418
P APNS PPT +++L +P+ P S +SPS + +NS+ +PP +H
Sbjct: 714 PLAPNS----NPPTKDSLLPRSPVKPMDSI---TSPSIAELHNSVTNCDPPLRLTKPSHF 766
Query: 419 ITRSQNNIFKPKKFFYA-SNHPLPENLEPSNVRQAMQHPHWRQAISEE 465
NI +K ++ SN P P NL ++ + + + + S E
Sbjct: 767 -----TNIKPVEKLDHSRSNTPSPLNLSSTSSKNSHSSSYTPNSFSSE 809
>ATX2_MOUSE (O70305) Ataxin-2 (Spinocerebellar ataxia type 2 protein
homolog)
Length = 1285
Score = 40.4 bits (93), Expect = 0.023
Identities = 34/128 (26%), Positives = 47/128 (36%), Gaps = 17/128 (13%)
Query: 265 SGATHHLTNNLDDLHITNPYHGSDKIIIGDGNT----LPISHVDPKTNKIYTSRHVVF-- 318
S HH LH+ +P S G T P S+ + ++ VF
Sbjct: 1142 SPVQHHQHQAAQALHLASPQQQSAIYHAGLAPTPPSMTPASNTQSPQSSFPAAQQTVFTI 1201
Query: 319 HDNHFP--YPTINQNAHAANLPINISLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAI 376
H +H Y T AH + ++P H P AP LM T PP P+A
Sbjct: 1202 HPSHVQPAYTTPPHMAHVPQAHVQSGMVPSH---------PTAHAPMMLMTTQPPGPKAA 1252
Query: 377 LQEAPLSP 384
L ++ L P
Sbjct: 1253 LAQSALQP 1260
>VRP1_YEAST (P37370) Verprolin
Length = 817
Score = 39.3 bits (90), Expect = 0.051
Identities = 26/94 (27%), Positives = 43/94 (45%), Gaps = 8/94 (8%)
Query: 340 NISLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPL----SPSTSSGNESSPS 395
N + +PP+ S+ PPP P +T+ + + APL PS ++ S+P
Sbjct: 383 NTTSVPPNKASSMPAPPPPPPPPPGAFSTSSALSASSIPLAPLPPPPPPSVATSVPSAPP 442
Query: 396 PSPTNNSIALTNPPADSNATNHIITRSQNNIFKP 429
P PT TN P+ S+ + I + S ++ P
Sbjct: 443 PPPT----LTTNKPSASSKQSKISSSSSSSAVTP 472
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 39.3 bits (90), Expect = 0.051
Identities = 21/64 (32%), Positives = 29/64 (44%)
Query: 345 PPHTILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPSPSPTNNSIA 404
PP + +PP AP S + PP+P SPS S SPSPSP+ +
Sbjct: 310 PPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSPSPSPSP 369
Query: 405 LTNP 408
+ +P
Sbjct: 370 IPSP 373
Score = 39.3 bits (90), Expect = 0.051
Identities = 26/75 (34%), Positives = 37/75 (48%), Gaps = 2/75 (2%)
Query: 338 PINISLIPPH-TILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPS- 395
P+ S PP T S +P +PP AP S PP+P P+ PS + + SP+
Sbjct: 166 PVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAP 225
Query: 396 PSPTNNSIALTNPPA 410
PSP + + +PPA
Sbjct: 226 PSPPSPAPPSPSPPA 240
Score = 37.0 bits (84), Expect = 0.25
Identities = 23/70 (32%), Positives = 29/70 (40%), Gaps = 2/70 (2%)
Query: 345 PPHTILSLNPTIPP--PQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPSPSPTNNS 402
PP N +PP P P S TPPTP + +P+ PS + S PSP +
Sbjct: 275 PPRPPFPANTPMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSP 334
Query: 403 IALTNPPADS 412
PP S
Sbjct: 335 PPSPAPPTPS 344
Score = 36.2 bits (82), Expect = 0.43
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query: 354 PTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSP-SPSPTNNSIALTNPPA 410
P +PP AP S PP+P + +P PS + + S P PSP S A PP+
Sbjct: 144 PPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPS 201
Score = 36.2 bits (82), Expect = 0.43
Identities = 24/73 (32%), Positives = 34/73 (45%), Gaps = 6/73 (8%)
Query: 338 PINISLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPSPS 397
P+ S PP S P +PP AP S + PP+P + +P P+ S PSP+
Sbjct: 197 PVPPSPAPP----SPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPS--PVPPSPA 250
Query: 398 PTNNSIALTNPPA 410
P + + PPA
Sbjct: 251 PPSPAPPSPKPPA 263
Score = 34.3 bits (77), Expect = 1.6
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query: 351 SLNPTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPS-PSPTNNSIALTNPP 409
S +P PP +P S PP+P P+ PS S SP+ PSPT S + PP
Sbjct: 128 SPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPP 187
Query: 410 A 410
+
Sbjct: 188 S 188
Score = 33.1 bits (74), Expect = 3.7
Identities = 23/55 (41%), Positives = 29/55 (51%), Gaps = 2/55 (3%)
Query: 357 PPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPS-PSPTNNSIALTNPPA 410
PP AP S + PP+P A AP SP+ S SP+ PSP S A +PP+
Sbjct: 70 PPSPAPPSPPSPAPPSP-APPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPS 123
Score = 33.1 bits (74), Expect = 3.7
Identities = 23/72 (31%), Positives = 30/72 (40%), Gaps = 3/72 (4%)
Query: 342 SLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPL---SPSTSSGNESSPSPSP 398
S +PP PP AP+ + PPTP +P SPS S SPSP P
Sbjct: 312 SPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSPSPSPSPIP 371
Query: 399 TNNSIALTNPPA 410
+ + +P A
Sbjct: 372 SPSPKPSPSPVA 383
Score = 33.1 bits (74), Expect = 3.7
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Query: 338 PINISLIPPHTILSLNPTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSP-SP 396
P+ S PP S +P +PP +P + PP+P P+ PS + + + P P
Sbjct: 145 PLPPSPAPP----SPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPP 200
Query: 397 SPTNNSIALTNPPA 410
SP S A PP+
Sbjct: 201 SPAPPSPAPPVPPS 214
Score = 32.3 bits (72), Expect = 6.2
Identities = 19/63 (30%), Positives = 26/63 (41%), Gaps = 8/63 (12%)
Query: 345 PPHTILSLNPTIPPPQAPNSL--------MATTPPTPEAILQEAPLSPSTSSGNESSPSP 396
PP PT P P P+ + + PP+P +P P+ S SPSP
Sbjct: 294 PPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSP 353
Query: 397 SPT 399
SP+
Sbjct: 354 SPS 356
Score = 32.3 bits (72), Expect = 6.2
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 3/48 (6%)
Query: 354 PTIPPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNES---SPSPSP 398
P PP AP S PP+P PL PS + + S PSPSP
Sbjct: 118 PPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSP 165
>G112_HUMAN (Q8IZF6) G-protein coupled receptor GPR112
Length = 2799
Score = 38.9 bits (89), Expect = 0.067
Identities = 25/92 (27%), Positives = 46/92 (49%), Gaps = 1/92 (1%)
Query: 357 PPPQAPNSLMATTPPTPEAILQEAPLSPSTSSGNESSPSPSPTNNSIALTNPPADSNATN 416
P ++ ++L A TP T E I+ ++ S S G ++S + T++S ++NP + +
Sbjct: 1314 PEKESTSALPAYTPRTVEMIVNSTYVTHSVSYGQDTSFVDTTTSSSTRISNPMDINTTFS 1373
Query: 417 HIIT-RSQNNIFKPKKFFYASNHPLPENLEPS 447
H+ + R+Q + F S PE+L S
Sbjct: 1374 HLHSLRTQPEVTSVASFISESTQTFPESLSLS 1405
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,777,117
Number of Sequences: 164201
Number of extensions: 5470861
Number of successful extensions: 20991
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 187
Number of HSP's that attempted gapping in prelim test: 19141
Number of HSP's gapped (non-prelim): 1129
length of query: 964
length of database: 59,974,054
effective HSP length: 120
effective length of query: 844
effective length of database: 40,269,934
effective search space: 33987824296
effective search space used: 33987824296
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146573.8


