

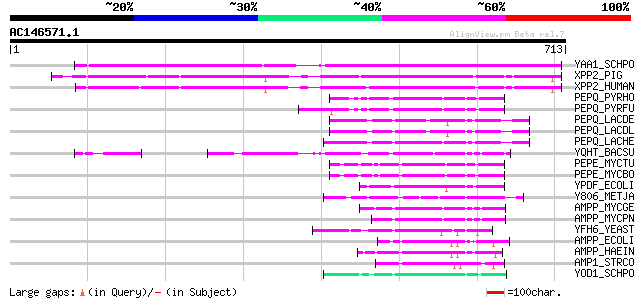
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.1 - phase: 0
(713 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YAA1_SCHPO (Q09795) Probable peptidase C22G7.01c (EC 3.4.-.-) 514 e-145
XPP2_PIG (Q95333) Xaa-Pro aminopeptidase 2 precursor (EC 3.4.11.... 419 e-116
XPP2_HUMAN (O43895) Xaa-Pro aminopeptidase 2 precursor (EC 3.4.1... 404 e-112
PEPQ_PYRHO (O58885) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dip... 94 2e-18
PEPQ_PYRFU (P81535) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dip... 93 3e-18
PEPQ_LACDE (Q9S6S1) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dip... 91 1e-17
PEPQ_LACDL (P46545) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dip... 90 2e-17
PEPQ_LACHE (O84913) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro dip... 85 8e-16
YQHT_BACSU (P54518) Putative peptidase yqhT (EC 3.4.-.-) 84 1e-15
PEPE_MYCTU (P65810) Probable dipeptidase pepE (EC 3.4.13.-) 70 1e-11
PEPE_MYCBO (P65811) Probable dipeptidase pepE (EC 3.4.13.-) 70 1e-11
YPDF_ECOLI (P76524) Putative peptidase ypdF (EC 3.4.-.-) 70 2e-11
Y806_METJA (Q58216) Hypothetical peptidase MJ0806 (EC 3.4.-.-) 69 4e-11
AMPP_MYCGE (P47566) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9... 68 1e-10
AMPP_MYCPN (P75313) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9... 67 2e-10
YFH6_YEAST (P43590) Hypothetical 61.8 kDa peptidase in MPR1-GCN2... 52 4e-06
AMPP_ECOLI (P15034) Xaa-Pro aminopeptidase (EC 3.4.11.9) (X-Pro ... 50 2e-05
AMPP_HAEIN (P44881) Xaa-Pro aminopeptidase (EC 3.4.11.9) (X-Pro ... 48 1e-04
AMP1_STRCO (Q05813) Xaa-Pro aminopeptidase I (EC 3.4.11.9) (X-Pr... 46 3e-04
YOD1_SCHPO (Q9UUD8) Probable peptidase C18A7.01 (EC 3.4.-.-) 45 5e-04
>YAA1_SCHPO (Q09795) Probable peptidase C22G7.01c (EC 3.4.-.-)
Length = 598
Score = 514 bits (1325), Expect = e-145
Identities = 275/626 (43%), Positives = 380/626 (59%), Gaps = 36/626 (5%)
Query: 84 KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAA 143
+L LR L + ++ Y++PS+DAH SE+ + ARR +IS F GS G AV+ AA
Sbjct: 8 RLNKLRELMKERGYTL--YVVPSEDAHSSEYTCDADARRAFISGFDGSAGCAVIGETSAA 65
Query: 144 LWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEE 203
L+TDGRYF QA +QL+ NW LM+ G GVPT E+ ++ +VGID L T AA+
Sbjct: 66 LFTDGRYFNQASQQLDENWTLMKQGFTGVPTWEEYCTQMTKCNEKVGIDSSLITFPAAKA 125
Query: 204 LKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSEL 263
L+ + K+ ++ + NLVD +W +RP+ P + + V +KYAGL V KL +LR +
Sbjct: 126 LRESLFLKSGAVLVGDHDNLVDIVWGASRPKEPLEKLIVQEIKYAGLGVDEKLHNLREAM 185
Query: 264 VQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDH 323
+ A +V+ LDE+AWL NLRG+D+P++PV +AY +V +D A L++D KVT EV H
Sbjct: 186 KEQKIEAFVVSMLDEVAWLYNLRGADVPYNPVFFAYSLVTLDEAFLYVDERKVTPEVSKH 245
Query: 324 LKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKT 383
L ++I PY+ + S+ +N + TS A K
Sbjct: 246 LD-GFVKILPYDRVFSDAKNSNLTRIGISSKTSWCIATSFGETKV--------------- 289
Query: 384 RSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKE 443
+PI SP+S AK IKN+ ELKGM+ECH+RD AL +++ WL++
Sbjct: 290 -------------MPIL----SPISQAKGIKNDAELKGMKECHIRDGCALVEYFAWLDEY 332
Query: 444 ITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANK 503
+ + + E + + KL +FR K F+ SF+TIS +GPNGA+IHY P + +D K
Sbjct: 333 LNSGNKINEFDAATKLEQFRRKNNLFMGLSFETISSTGPNGAVIHYSPPATGSAIIDPTK 392
Query: 504 LFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDA 563
++L DSGAQY DGTTD+TRT HFG+P+ E++ T L+GHIAL VFP+ T G+++D
Sbjct: 393 IYLCDSGAQYKDGTTDVTRTWHFGEPSEFERQTATLALKGHIALANIVFPKGTTGYMIDV 452
Query: 564 FARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYR-YGNLTPLVNGMIVSNEPGYYED 622
AR +LWK GLDY HGTGHGVG+ LNVHE P GI R N PL GM+ SNEPG+YED
Sbjct: 453 LARQYLWKYGLDYLHGTGHGVGSFLNVHELPVGIGSREVFNSAPLQAGMVTSNEPGFYED 512
Query: 623 HAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYH 682
FG R+EN +Y+ V T NRF G YLG + LT P KL+D SLLS E+ +LN YH
Sbjct: 513 GHFGYRVENCVYITEVNTENRFAGRTYLGLKDLTLAPHCQKLIDPSLLSPEEVKYLNEYH 572
Query: 683 SVVWEKVSPLLDGSARQWLWNNTQPI 708
S V+ +SP+L SA++WL +T PI
Sbjct: 573 SEVYTTLSPMLSVSAKKWLSKHTSPI 598
>XPP2_PIG (Q95333) Xaa-Pro aminopeptidase 2 precursor (EC 3.4.11.9)
(X-Pro aminopeptidase 2) (Membrane-bound aminopeptidase
P) (Membrane-bound APP) (Membrane-bound AmP) (mAmP)
(Aminoacylproline aminopeptidase)
Length = 673
Score = 419 bits (1077), Expect = e-116
Identities = 255/666 (38%), Positives = 369/666 (55%), Gaps = 61/666 (9%)
Query: 54 IRNCTTSNSISAKPSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSE 113
+RNC+TS + ++ +LTALR ++S AYIIP DAH SE
Sbjct: 32 VRNCSTSPPYLPVTAV----------NTTAQLTALREQMLTQNLS--AYIIPDTDAHMSE 79
Query: 114 FIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVP 173
+I + RR +I+ F GS G AVVT KAALWTD RY+ QAE+Q++ NW L + + G
Sbjct: 80 YIGECDQRRAWITGFIGSAGIAVVTERKAALWTDSRYWTQAERQMDCNWELHKEVSTGHI 139
Query: 174 TTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARP 233
T WL + G RVG DPFLF+ D+ E + + ELV + NLVD +W RP
Sbjct: 140 VT--WLLTEIPVGGRVGFDPFLFSIDSWESYDVALQDADRELVSI-TVNLVDLVWGSERP 196
Query: 234 EPPNKPVRVHGLKYAGLDVASKLSSLRSELVQ--AGSSAIIVTALDEIAWLLNLRGSDIP 291
PN P+ +AG K+S++RS++ + +A++++ALDE AWL NLR SDIP
Sbjct: 197 PLPNAPIYALQEAFAGSTWQEKVSNIRSQMQKHHERPTAVLLSALDETAWLFNLRSSDIP 256
Query: 292 HSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKA-----NIEIRPYNSIVSEIENLAA 346
++P Y+Y ++ +LF + S+ + E +L + +++ Y+ I I+ +
Sbjct: 257 YNPFFYSYTLLTDSSIRLFANKSRFSSETLQYLNSSCNSSMCVQLEDYSQIRDSIQAYTS 316
Query: 347 RGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSP 406
+W+ T + Y YE K + + SP
Sbjct: 317 GDVKIWIGTRYTS--------------YGLYEVIPKEK--------------LVEDDYSP 348
Query: 407 VSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQ 466
V + KA+KN E ++ H+RDA A+ ++ WLEK + + E + ++ EFR ++
Sbjct: 349 VMITKAVKNSREQALLKASHVRDAVAVIRYLAWLEKNVPTGTV-DEFSGAKRVEEFRGEE 407
Query: 467 AGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHF 526
F SF+TIS SG N A+ HY P + +++++LLDSG QY DGTTDITRTVH+
Sbjct: 408 EFFSGPSFETISASGLNAALAHYSPTKELHRKLSSDEMYLLDSGGQYWDGTTDITRTVHW 467
Query: 527 GKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGA 586
G P+ +KE +TRVL G+I L + VFP T G V++AFAR LW VGL+Y HGTGHG+G
Sbjct: 468 GTPSAFQKEAYTRVLIGNIDLSRLVFPAATSGRVVEAFARKALWDVGLNYGHGTGHGIGN 527
Query: 587 ALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGG 646
L VHE P G ++YGN+ P+ GM S EPGYY+D FGIR+E++ V VE ++ G
Sbjct: 528 FLCVHEWPVG--FQYGNI-PMAEGMFTSIEPGYYQDGEFGIRLEDVALV--VEAKTKYPG 582
Query: 647 IQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDG----SARQWLW 702
YL FE ++ VP KL+DVSLLS ++ +LN Y+ + EKV P L WL
Sbjct: 583 -TYLTFEVVSLVPYDRKLIDVSLLSPEQLQYLNRYYQAIREKVGPELQRRGLLEELSWLQ 641
Query: 703 NNTQPI 708
+T+P+
Sbjct: 642 RHTEPL 647
>XPP2_HUMAN (O43895) Xaa-Pro aminopeptidase 2 precursor (EC
3.4.11.9) (X-Pro aminopeptidase 2) (Membrane-bound
aminopeptidase P) (Membrane-bound APP) (Membrane-bound
AmP) (mAmP) (Aminoacylproline aminopeptidase)
Length = 674
Score = 404 bits (1039), Expect = e-112
Identities = 242/637 (37%), Positives = 349/637 (53%), Gaps = 55/637 (8%)
Query: 85 LTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAAL 144
LTALR+ ++S AYIIP DAH +E+I Q RR +I+ FTGS GTAVVT KAA+
Sbjct: 54 LTALRQQMQTQNLS--AYIIPGTDAHMNEYIGQHDERRAWITGFTGSAGTAVVTMKKAAV 111
Query: 145 WTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEEL 204
WTD RY+ QAE+Q++ NW L + G WL + G RVG DPFL + D E
Sbjct: 112 WTDSRYWTQAERQMDCNWELHK--EVGTTPIVTWLLTEIPAGGRVGFDPFLLSIDTWESY 169
Query: 205 KHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELV 264
+ N +LV + +NLVD +W RP PN+P+ + G K+S +RS++
Sbjct: 170 DLALQGSNRQLVSI-TTNLVDLVWGSERPPVPNQPIYALQEAFTGSTWQEKVSGVRSQMQ 228
Query: 265 --QAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDD 322
Q +A++++AL+E AWL NLR SDIP++P Y+Y ++ +LF + S+ + E
Sbjct: 229 KHQKVPTAVLLSALEETAWLFNLRASDIPYNPFFYSYTLLTDSSIRLFANKSRFSSETLS 288
Query: 323 HLKKA-----NIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNY 377
+L + ++I Y+ + I+ + +W+ TS Y Y
Sbjct: 289 YLNSSCTGPMCVQIEDYSQVRDSIQAYSLGDVRIWIGTSYT--------------MYGIY 334
Query: 378 ESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFW 437
E K + + SPV + KA+KN E ++ H+RDA A+ ++
Sbjct: 335 EMIPKEK--------------LVTDTYSPVMMTKAVKNSKEQALLKASHVRDAVAVIRYL 380
Query: 438 DWLEKEITNDRI--LTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGS 495
WLEK + + + E+ DK FR ++ SF+TIS SG N A+ HY P
Sbjct: 381 VWLEKNVPKGTVDEFSGAEIVDK---FRGEEQFSSGPSFETISASGLNAALAHYSPTKEL 437
Query: 496 CSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPED 555
+ +++++LLDSG QY DGTTDITRTVH+G P+ +KE +TRVL G+I L + +FP
Sbjct: 438 NRKLSSDEMYLLDSGGQYWDGTTDITRTVHWGTPSAFQKEAYTRVLIGNIDLSRLIFPAA 497
Query: 556 TPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSN 615
T G +++AFAR LW GL+Y HGTGHG+G L VHE P G N + GM S
Sbjct: 498 TSGRMVEAFARRALWDAGLNYGHGTGHGIGNFLCVHEWPVGFQ---SNNIAMAKGMFTSI 554
Query: 616 EPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEI 675
EPGYY+D FGIR+E++ V VE ++ G YL FE +++VP L+DVSLLS +
Sbjct: 555 EPGYYKDGEFGIRLEDVALV--VEAKTKYPG-SYLTFEVVSFVPYDRNLIDVSLLSPEHL 611
Query: 676 DWLNNYHSVVWEKVSPLLDG----SARQWLWNNTQPI 708
+LN Y+ + EKV P L +WL +T+P+
Sbjct: 612 QYLNRYYQTIREKVGPELQRRQLLEEFEWLQQHTEPL 648
>PEPQ_PYRHO (O58885) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro
dipeptidase) (Proline dipeptidase) (Prolidase)
(Imidodipeptidase)
Length = 351
Score = 93.6 bits (231), Expect = 2e-18
Identities = 75/228 (32%), Positives = 116/228 (49%), Gaps = 20/228 (8%)
Query: 411 KAIKNETELKGMQE-CHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGF 469
+ IK+E E+K +++ C + D A +A +EIT + E EV+ K+ E+ K G
Sbjct: 125 RIIKSEKEIKIIEKACEIADKAVMAAI-----EEITEGK--KEREVAAKV-EYLMKMNGA 176
Query: 470 LDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKP 529
+FDTI SG A+ H S ++ L ++D GA Y +DITRT+ G P
Sbjct: 177 EKPAFDTIIASGYRSALPH---GVASDKRIERGDLVVIDLGALYQHYNSDITRTIVVGSP 233
Query: 530 TTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTGHGVGAA 587
++KE + VL+ ++ P T LD+ AR+ + + G + H GHGVG
Sbjct: 234 NEKQKEIYEIVLEAQKKAVESAKPGIT-AKELDSIARNIIAEYGYGEYFNHSLGHGVG-- 290
Query: 588 LNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYV 635
L VHE P+ Y + T L GM+++ EPG Y G+RIE+ + +
Sbjct: 291 LEVHEWPRVSQY---DETVLREGMVITIEPGIYIPKIGGVRIEDTILI 335
>PEPQ_PYRFU (P81535) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro
dipeptidase) (Proline dipeptidase) (Prolidase)
(Imidodipeptidase)
Length = 348
Score = 92.8 bits (229), Expect = 3e-18
Identities = 82/276 (29%), Positives = 132/276 (47%), Gaps = 29/276 (10%)
Query: 372 RYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKA---------IKNETELKGM 422
++ + YE T + G +G+++ S V KS+ K IK + E++ +
Sbjct: 74 KFDEIYEILKNTETLGIEGTLSYSMVENFKEKSNVKEFKKIDDVIKDLRIIKTKEEIEII 133
Query: 423 QE-CHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSG 481
++ C + D A +A +EIT + E EV+ K+ E+ K G +FDTI SG
Sbjct: 134 EKACEIADKAVMAAI-----EEITEGK--REREVAAKV-EYLMKMNGAEKPAFDTIIASG 185
Query: 482 PNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVL 541
A+ H S ++ L ++D GA Y +DITRT+ G P +++E + VL
Sbjct: 186 HRSALPH---GVASDKRIERGDLVVIDLGALYNHYNSDITRTIVVGSPNEKQREIYEIVL 242
Query: 542 QGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTGHGVGAALNVHEGPQGISY 599
+ +A P T LD+ AR + + G + H GHGVG L +HE P+ Y
Sbjct: 243 EAQKRAVEAAKPGMT-AKELDSIAREIIKEYGYGDYFIHSLGHGVG--LEIHEWPRISQY 299
Query: 600 RYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYV 635
+ T L GM+++ EPG Y G+RIE+ + +
Sbjct: 300 ---DETVLKEGMVITIEPGIYIPKLGGVRIEDTVLI 332
>PEPQ_LACDE (Q9S6S1) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro
dipeptidase) (Proline dipeptidase) (Prolidase)
(Imidodipeptidase)
Length = 368
Score = 90.5 bits (223), Expect = 1e-17
Identities = 78/262 (29%), Positives = 118/262 (44%), Gaps = 35/262 (13%)
Query: 411 KAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFL 470
+ K E+EL +++ A ++ L +T +++++E KL Q G +
Sbjct: 135 RLFKTESELVKLRKAGEEADFAFQIGFEALRNGVTERAVVSQIEYQLKL------QKGVM 188
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPT 530
TSFDTI +G N A H P S +TV N+L L D G + +D +RTV +G+PT
Sbjct: 189 QTSFDTIVQAGKNAANPHQGP---SMNTVQPNELVLFDLGTMHEGYASDSSRTVAYGEPT 245
Query: 531 TREKECFTRVLQGHIALDQAVFPEDTPGFV---LDAFARSFLWKVGLD--YRHGTGHGVG 585
+ +E + + + QA PG LD AR + G + H GHG+G
Sbjct: 246 DKMRE----IYEVNRTAQQAAIDAAKPGMTASELDGVARKIITDAGYGEYFIHRLGHGIG 301
Query: 586 AALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFG 645
+ VHE P S GN L GM S EPG Y G+RIE+ G
Sbjct: 302 --MEVHEFP---SIANGNDVVLEEGMCFSIEPGIYIPGFAGVRIEDC------------G 344
Query: 646 GIQYLGFEKLTYVPIQIKLVDV 667
+ GF+ T+ ++K++ V
Sbjct: 345 VLTKEGFKPFTHTSKELKVLPV 366
>PEPQ_LACDL (P46545) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro
dipeptidase) (Proline dipeptidase) (Prolidase)
(Imidodipeptidase)
Length = 368
Score = 90.1 bits (222), Expect = 2e-17
Identities = 78/262 (29%), Positives = 118/262 (44%), Gaps = 35/262 (13%)
Query: 411 KAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFL 470
+ K E+EL +++ A ++ L +T +++++E KL Q G +
Sbjct: 135 RLFKTESELVKLRKAGEEADFAFQIGFEALRNGVTERAVVSQIEYQLKL------QKGVM 188
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPT 530
TSFDTI +G N A H P S +TV N+L L D G + +D +RTV +G+PT
Sbjct: 189 QTSFDTIVQAGKNAANPHQGP---SMNTVQPNELVLFDLGTMHEGYASDSSRTVAYGEPT 245
Query: 531 TREKECFTRVLQGHIALDQAVFPEDTPGFV---LDAFARSFLWKVGLD--YRHGTGHGVG 585
+ +E + + + QA PG LD AR + G + H GHG+G
Sbjct: 246 DKMRE----IYEVNRTAQQAAIDAAKPGMTASELDGVARKIITDAGYGEYFIHRLGHGIG 301
Query: 586 AALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFG 645
+ VHE P S GN L GM S EPG Y G+RIE+ G
Sbjct: 302 --MEVHEFP---SIANGNDVVLEEGMCFSIEPGIYIPGFAGVRIEDC------------G 344
Query: 646 GIQYLGFEKLTYVPIQIKLVDV 667
+ GF+ T+ ++K++ V
Sbjct: 345 VLTKDGFKPFTHTSKELKVLPV 366
>PEPQ_LACHE (O84913) Xaa-Pro dipeptidase (EC 3.4.13.9) (X-Pro
dipeptidase) (Proline dipeptidase) (Prolidase)
(Imidodipeptidase)
Length = 368
Score = 84.7 bits (208), Expect = 8e-16
Identities = 76/266 (28%), Positives = 117/266 (43%), Gaps = 29/266 (10%)
Query: 404 SSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFR 463
SS + + K E+K +Q A +D + +T I +++ K+
Sbjct: 128 SSFIERLRLYKTPEEIKKLQGAGAEADFAFKIGFDAIRTGVTERSIAGQIDYQLKI---- 183
Query: 464 SKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRT 523
Q G + SF+TI +G N A H P + +TV N+L L D G + +D +RT
Sbjct: 184 --QKGVMHESFETIVQAGKNAANPHLGP---TMNTVQPNELVLFDLGTMHDGYASDSSRT 238
Query: 524 VHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTG 581
V +G P+ +++E + + A +A P T LD+ AR + K G + H G
Sbjct: 239 VAYGTPSDKQREIYEVDREAQQAAIEAAKPGIT-AEELDSVARDIITKAGYGEYFIHRLG 297
Query: 582 HGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETP 641
HG+G NVHE P S GN L GM S EPG Y G+RIE+
Sbjct: 298 HGIGK--NVHEYP---SIVQGNDLVLEEGMCFSIEPGIYIPGFAGVRIEDC--------- 343
Query: 642 NRFGGIQYLGFEKLTYVPIQIKLVDV 667
G + GF+ T+ +K++ +
Sbjct: 344 ---GVVTKDGFKTFTHTDKDLKIIPI 366
>YQHT_BACSU (P54518) Putative peptidase yqhT (EC 3.4.-.-)
Length = 353
Score = 84.3 bits (207), Expect = 1e-15
Identities = 102/392 (26%), Positives = 159/392 (40%), Gaps = 55/392 (14%)
Query: 255 KLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNS 314
KL LR+ Q G +++T+ + ++ GS A L V FI +
Sbjct: 2 KLEKLRNLFGQLGIDGMLITSNTNVRYMTGFTGS---------AGLAVISGDKAAFITDF 52
Query: 315 KVTEEVDDHLKKANIEIRPYNSIVSEIENLAARG-SSLWLDTSSVNAAIVNAYKAACDRY 373
+ TE+ +K I + I + + + + G L + +S+ +Y A
Sbjct: 53 RYTEQAKVQVKGFEIIEHGGSLIQTTADTVESFGIKRLGFEQNSMTYGTYASYSAVIS-- 110
Query: 374 YQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAAL 433
D + VP+A V + IK+ E+K ++E A
Sbjct: 111 ---------------DAEL----VPVA----ESVEKLRLIKSSEEIKILEEAAKIADDAF 147
Query: 434 AQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEP 493
+++ I+ + E LEF + G +SFD I SG ++ H
Sbjct: 148 RHILTFMKPGISEIAVANE-------LEFYMRSQGADSSSFDMIVASGLRSSLPH---GV 197
Query: 494 GSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFP 553
S +++ L LD GA Y +DITRTV G+P+ + KE + V AL A
Sbjct: 198 ASDKLIESGDLVTLDFGAYYKGYCSDITRTVAVGQPSDQLKEIYQVVFDAQ-ALGVAHIK 256
Query: 554 EDTPGFVLDAFARSFLWKVGLD--YRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGM 611
G DA R + G + H TGHG+G + VHE P G+S R + L GM
Sbjct: 257 PGMTGKEADALTRDHIAAKGYGDYFGHSTGHGLG--MEVHESP-GLSVRSSAI--LEPGM 311
Query: 612 IVSNEPGYYEDHAFGIRIENLLYVRNVETPNR 643
+V+ EPG Y G+RIE+ + + E NR
Sbjct: 312 VVTVEPGIYIPETGGVRIEDDIVI--TENGNR 341
Score = 44.7 bits (104), Expect = 9e-04
Identities = 31/86 (36%), Positives = 43/86 (49%), Gaps = 14/86 (16%)
Query: 84 KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAA 143
KL LR LF + + ID +I S +Y++ FTGS G AV++ DKAA
Sbjct: 2 KLEKLRNLFGQ--LGIDGMLITSN------------TNVRYMTGFTGSAGLAVISGDKAA 47
Query: 144 LWTDGRYFLQAEKQLNSNWILMRAGN 169
TD RY QA+ Q+ I+ G+
Sbjct: 48 FITDFRYTEQAKVQVKGFEIIEHGGS 73
>PEPE_MYCTU (P65810) Probable dipeptidase pepE (EC 3.4.13.-)
Length = 375
Score = 70.5 bits (171), Expect = 1e-11
Identities = 71/228 (31%), Positives = 112/228 (48%), Gaps = 20/228 (8%)
Query: 411 KAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFL 470
+ +K ETE+ +++ AA+ + + + + R TE +V+ + E + G
Sbjct: 144 RMVKEETEIDALRKA----GAAIDRVHARVPEFLVPGR--TEADVAADIAEAIVAE-GHS 196
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGT-TDITRTVHFGKP 529
+ +F I GSGP+GA H+ D + ++D G Y G +D TRT G+P
Sbjct: 197 EVAF-VIVGSGPHGADPHHGYSDRELREGD---IVVVDIGGTYGPGYHSDSTRTYSIGEP 252
Query: 530 TTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTGHGVGAA 587
+ + ++ + + A +A+ P T V DA AR L + GL + H TGHG+G
Sbjct: 253 DSDVAQSYSMLQRAQRAAFEAIRPGVTAEQV-DAAARDVLAEAGLAEYFVHRTGHGIG-- 309
Query: 588 LNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYV 635
L VHE P ++ GN LV GM S EPG Y +G RIE+++ V
Sbjct: 310 LCVHEEPYIVA---GNDLVLVPGMAFSIEPGIYFPGRWGARIEDIVIV 354
>PEPE_MYCBO (P65811) Probable dipeptidase pepE (EC 3.4.13.-)
Length = 375
Score = 70.5 bits (171), Expect = 1e-11
Identities = 71/228 (31%), Positives = 112/228 (48%), Gaps = 20/228 (8%)
Query: 411 KAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFL 470
+ +K ETE+ +++ AA+ + + + + R TE +V+ + E + G
Sbjct: 144 RMVKEETEIDALRKA----GAAIDRVHARVPEFLVPGR--TEADVAADIAEAIVAE-GHS 196
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGT-TDITRTVHFGKP 529
+ +F I GSGP+GA H+ D + ++D G Y G +D TRT G+P
Sbjct: 197 EVAF-VIVGSGPHGADPHHGYSDRELREGD---IVVVDIGGTYGPGYHSDSTRTYSIGEP 252
Query: 530 TTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTGHGVGAA 587
+ + ++ + + A +A+ P T V DA AR L + GL + H TGHG+G
Sbjct: 253 DSDVAQSYSMLQRAQRAAFEAIRPGVTAEQV-DAAARDVLAEAGLAEYFVHRTGHGIG-- 309
Query: 588 LNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYV 635
L VHE P ++ GN LV GM S EPG Y +G RIE+++ V
Sbjct: 310 LCVHEEPYIVA---GNDLVLVPGMAFSIEPGIYFPGRWGARIEDIVIV 354
>YPDF_ECOLI (P76524) Putative peptidase ypdF (EC 3.4.-.-)
Length = 361
Score = 70.1 bits (170), Expect = 2e-11
Identities = 62/192 (32%), Positives = 93/192 (48%), Gaps = 15/192 (7%)
Query: 450 LTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDS 509
++E E++ +L E+ +Q G SFDTI SG GA+ H K S V A + LD
Sbjct: 158 MSEREIAAEL-EWFMRQQGAEKASFDTIVASGWRGALPHGK---ASDKIVAAGEFVTLDF 213
Query: 510 GAQYVDGTTDITRTVHF-GKPTTREKECFTRVLQGHIALDQAVFPEDTPGF---VLDAFA 565
GA Y +D+TRT+ G+ + E V Q + A PG +D A
Sbjct: 214 GALYQGYCSDMTRTLLVNGEGVSAESHLLFNVYQIVLQAQLAAISAIRPGVRCQQVDDAA 273
Query: 566 RSFLWKVGLD--YRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDH 623
R + + G + H TGH +G + VHE P+ + + T L GM+++ EPG Y
Sbjct: 274 RRVITEAGYGDYFGHNTGHAIG--IEVHEDPR---FSPRDTTTLQPGMLLTVEPGIYLPG 328
Query: 624 AFGIRIENLLYV 635
G+RIE+++ V
Sbjct: 329 QGGVRIEDVVLV 340
>Y806_METJA (Q58216) Hypothetical peptidase MJ0806 (EC 3.4.-.-)
Length = 347
Score = 68.9 bits (167), Expect = 4e-11
Identities = 71/260 (27%), Positives = 117/260 (44%), Gaps = 31/260 (11%)
Query: 404 SSPVSLAKAIKNETELKGMQEC-HLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEF 462
S + + IK++ E+K +++ + D A +W+ + + LTE E+ ++ E+
Sbjct: 113 SDKIKEMRMIKDKEEIKLIKKAAEISDKAI-----NWVLNNLDEVKNLTEYELVAEI-EY 166
Query: 463 RSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITR 522
K+ G + +FD+I SG + H P + + L+D GA Y +DITR
Sbjct: 167 IMKKHGSIKPAFDSIVVSGKKTSFPHALPTKDKIADI-----LLVDIGAVYEGYCSDITR 221
Query: 523 TVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGH 582
T K K+ + V + ++ + E +D R F + H GH
Sbjct: 222 TFLL-KDDEEMKKIYNLVYEAKKVAEEHL-KEGISAKQIDNIVREFFNDYKELFIHSLGH 279
Query: 583 GVGAALNVHEGPQGISYRYGNLTPLV--NGMIVSNEPGYYEDHAFGIRIENLLYVRNVET 640
GVG L VHE P+ +S + + ++ GM+V+ EPG Y FG+RIE+L V+
Sbjct: 280 GVG--LEVHEEPR-LSNKLKDDEDIILKEGMVVTIEPGLYLKDKFGVRIEDLYLVKK--- 333
Query: 641 PNRFGGIQYLGFEKLTYVPI 660
GFEKL+ I
Sbjct: 334 ---------NGFEKLSKAEI 344
>AMPP_MYCGE (P47566) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9)
(X-Pro aminopeptidase) (Aminopeptidase P) (APP)
(Aminoacylproline aminopeptidase)
Length = 354
Score = 67.8 bits (164), Expect = 1e-10
Identities = 62/191 (32%), Positives = 90/191 (46%), Gaps = 14/191 (7%)
Query: 450 LTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDS 509
+TE+ +S + KQ G SFD I +G NGA H+KP + + V D
Sbjct: 159 MTELFISQWITNELVKQGG-AKNSFDPIVATGKNGANPHHKP---TKTIVKEGDFITCDF 214
Query: 510 GAQYVDGTTDITRTVHFGKPTTREK--ECFTRVLQGHIALDQAVFPEDTPGFVLDAFARS 567
G Y +DITRT GK K + +V + ++A AV T G +D R
Sbjct: 215 GTIYNGYCSDITRTFLVGKKPKSAKLLSAYKKVEEANLAGINAVNTTLT-GSQVDKVCRD 273
Query: 568 FLWKVGLD--YRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAF 625
+ + H TGHGVG +++HE P +S Y L L +V+ EPG Y +
Sbjct: 274 IIENSEFKDFFVHSTGHGVG--IDIHEMP-NVSQSYNKL--LCENGVVTIEPGIYIPNLG 328
Query: 626 GIRIENLLYVR 636
GIRIE+++ V+
Sbjct: 329 GIRIEDMVLVK 339
Score = 32.3 bits (72), Expect = 4.5
Identities = 14/38 (36%), Positives = 24/38 (62%)
Query: 122 RKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLN 159
R +++ F S G ++T++KA L+ DGRY+ A +N
Sbjct: 32 RFWLTNFPSSAGWLIITSNKAKLFIDGRYYEAARNFIN 69
>AMPP_MYCPN (P75313) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9)
(X-Pro aminopeptidase) (Aminopeptidase P) (APP)
(Aminoacylproline aminopeptidase)
Length = 354
Score = 66.6 bits (161), Expect = 2e-10
Identities = 58/176 (32%), Positives = 84/176 (46%), Gaps = 13/176 (7%)
Query: 466 QAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVH 525
+AG SFD I +G NGA H+KP S V + D G Y +DITRT
Sbjct: 174 KAGGAKNSFDPIVATGKNGANPHHKP---SKLKVKSGDFVTCDFGTIYNGYCSDITRTFL 230
Query: 526 FGKPTTRE--KECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRHGTG 581
GK E + + +V + ++A A + T G +D R + + H TG
Sbjct: 231 VGKKPNNEVLLKAYKKVDEANMAGINAANTQLT-GAEVDKVCRDIIEASEFKDYFVHSTG 289
Query: 582 HGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRN 637
HGVG L++HE P +S Y L L +++ EPG Y GIRIE+++ V++
Sbjct: 290 HGVG--LDIHEMP-NVSTSYNKL--LCENAVITIEPGIYIPSVGGIRIEDMVLVKD 340
>YFH6_YEAST (P43590) Hypothetical 61.8 kDa peptidase in MPR1-GCN20
intergenic region (EC 3.4.-.-)
Length = 535
Score = 52.4 bits (124), Expect = 4e-06
Identities = 66/264 (25%), Positives = 109/264 (41%), Gaps = 46/264 (17%)
Query: 389 DGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQE-CHLRDAAALAQFWDWLEKEITND 447
D +IA S +P + + +AIK+ E++ +++ C + D + LA L E+
Sbjct: 207 DENIARSLIPSDPNFFYAMDETRAIKDWYEIESIRKACQISDKSHLAVM-SALPIELNEL 265
Query: 448 RILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLL 507
+I E E+ + + G +D I SGP +HY + + L+
Sbjct: 266 QIQAE-------FEYHATRQGGRSLGYDPICCSGPACGTLHYVK---NSEDIKGKHSILI 315
Query: 508 DSGAQYVDGTTDITRTV-HFGKPTTREKECFTRVLQGHIALDQAVFP----EDTPGFVLD 562
D+GA++ T+DITR GK T +E + VL + + P +D
Sbjct: 316 DAGAEWRQYTSDITRCFPTSGKFTAEHREVYETVLDMQNQAMERIKPGAKWDDLHALTHK 375
Query: 563 AFARSFLWKVGL------------------DYRHGTGHGVGAALNVHEGPQGISY----- 599
+ FL +G+ Y HG GH +G L+VH+ +Y
Sbjct: 376 VLIKHFL-SMGIFKKEFSEDEIFKRRASCAFYPHGLGHMLG--LDVHDVGGNPNYDDPDP 432
Query: 600 --RYGNL-TPLVNGMIVSNEPGYY 620
RY + PL M+++NEPG Y
Sbjct: 433 MFRYLRIRRPLKENMVITNEPGCY 456
>AMPP_ECOLI (P15034) Xaa-Pro aminopeptidase (EC 3.4.11.9) (X-Pro
aminopeptidase) (Aminopeptidase P II) (APP-II)
(Aminoacylproline aminopeptidase)
Length = 440
Score = 50.4 bits (119), Expect = 2e-05
Identities = 58/199 (29%), Positives = 85/199 (42%), Gaps = 37/199 (18%)
Query: 473 SFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHF-GKPTT 531
S++TI GSG NG I+HY C D + L L+D+G +Y DITRT GK T
Sbjct: 228 SYNTIVGSGENGCILHY--TENECEMRDGD-LVLIDAGCEYKGYAGDITRTFPVNGKFTQ 284
Query: 532 REKECFTRVLQGHIALDQAVFPEDTPGFVLDAFAR---SFLWKVGL-------------- 574
++E + VL+ + P + V R S L K+G+
Sbjct: 285 AQREIYDIVLESLETSLRLYRPGTSILEVTGEVVRIMVSGLVKLGILKGDVDELIAQNAH 344
Query: 575 --DYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYY---------EDH 623
+ HG H +G L+VH+ G+ Y L GM+++ EPG Y +
Sbjct: 345 RPFFMHGLSHWLG--LDVHD--VGV-YGQDRSRILEPGMVLTVEPGLYIAPDAEVPEQYR 399
Query: 624 AFGIRIENLLYVRNVETPN 642
GIRIE+ + + N
Sbjct: 400 GIGIRIEDDIVITETGNEN 418
>AMPP_HAEIN (P44881) Xaa-Pro aminopeptidase (EC 3.4.11.9) (X-Pro
aminopeptidase) (Aminopeptidase P II) (APP-II)
(Aminoacylproline aminopeptidase)
Length = 430
Score = 47.8 bits (112), Expect = 1e-04
Identities = 62/216 (28%), Positives = 92/216 (41%), Gaps = 40/216 (18%)
Query: 447 DRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFL 506
+R E+E SD L EF A F S+++I G N I+HY + D L L
Sbjct: 199 NRFEYEIE-SDILHEFNRHCARF--PSYNSIVAGGSNACILHYTENDRPLNDGD---LVL 252
Query: 507 LDSGAQYVDGTTDITRTVHF-GKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFA 565
+D+G ++ DITRT GK + ++E + VL+ + + P ++ D
Sbjct: 253 IDAGCEFAMYAGDITRTFPVNGKFSQPQREIYELVLKAQKRAIELLVPGNSIKQANDEVI 312
Query: 566 R---SFLWKVGL----------------DYRHGTGHGVGAALNVHEGPQGISYRYGNLTP 606
R L +G+ Y HG GH +G L+VH+ SY
Sbjct: 313 RIKTQGLVDLGILKGDVDTLIEQQAYRQFYMHGLGHWLG--LDVHDVG---SYGQDKQRI 367
Query: 607 LVNGMIVSNEPGYY--ED-------HAFGIRIENLL 633
L GM+++ EPG Y ED G+RIE+ L
Sbjct: 368 LEIGMVITVEPGIYISEDADVPEQYKGIGVRIEDNL 403
>AMP1_STRCO (Q05813) Xaa-Pro aminopeptidase I (EC 3.4.11.9) (X-Pro
aminopeptidase I) (Aminopeptidase P I) (APP) (PEPP I)
(Aminoacylproline aminopeptidase I)
Length = 490
Score = 46.2 bits (108), Expect = 3e-04
Identities = 54/196 (27%), Positives = 82/196 (41%), Gaps = 37/196 (18%)
Query: 471 DTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQ-YVDGTTDITRTVHF-GK 528
D + +I +GP+ +H+ G + D L LLD+G + + T D+TRT+ G
Sbjct: 273 DVGYGSICAAGPHACTLHWVRNDGPVRSGD---LLLLDAGVETHTYYTADVTRTLPISGT 329
Query: 529 PTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFL-----W-----------KV 572
+ +K+ + V A AV P DA R W ++
Sbjct: 330 YSELQKKIYDAVYDAQEAGIAAVRPGAKYRDFHDASQRVLAERLVEWGLVEGPVERVLEL 389
Query: 573 GLDYR---HGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYY--------- 620
GL R HGTGH +G ++ + SY G L P GM+++ EPG Y
Sbjct: 390 GLQRRWTLHGTGHMLGMDVHDCAAARVESYVDGTLEP---GMVLTVEPGLYFQADDLTVP 446
Query: 621 -EDHAFGIRIENLLYV 635
E G+RIE+ + V
Sbjct: 447 EEYRGIGVRIEDDILV 462
>YOD1_SCHPO (Q9UUD8) Probable peptidase C18A7.01 (EC 3.4.-.-)
Length = 451
Score = 45.4 bits (106), Expect = 5e-04
Identities = 62/240 (25%), Positives = 95/240 (38%), Gaps = 20/240 (8%)
Query: 404 SSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFR 463
S V+ + IK+ E+ M ++ AA+ ++ IT + EV + L +
Sbjct: 212 SPRVASLREIKSPAEVDIMSRVNIATVAAIRSVQPCIKPGITEKEL---AEVINMLFVY- 267
Query: 464 SKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRT 523
G L I G A+ H P S + ++ L+D G +D TRT
Sbjct: 268 ----GGLPVQESPIVLFGERAAMPHGGP---SNRRLKKSEFVLMDVGTTLFGYHSDCTRT 320
Query: 524 V--HFGKPTTREKECFTRVLQGHIA-LDQAVFPEDTPGFVLDAFARSFLWKVGLD--YRH 578
V H K T R ++ + V A + +T +D AR + G + H
Sbjct: 321 VLPHGQKMTERMEKLWNLVYDAQTAGIQMLSHLSNTSCAEVDLAARKVIKDAGYGEYFIH 380
Query: 579 GTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNV 638
GHG+G L HE G TP+ G + + EPG Y GIRIE+ + +V
Sbjct: 381 RLGHGLG--LEEHEQTYLNPANKG--TPVQKGNVFTVEPGIYIPDEIGIRIEDAVLASDV 436
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 86,523,324
Number of Sequences: 164201
Number of extensions: 3774679
Number of successful extensions: 9891
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 9818
Number of HSP's gapped (non-prelim): 60
length of query: 713
length of database: 59,974,054
effective HSP length: 117
effective length of query: 596
effective length of database: 40,762,537
effective search space: 24294472052
effective search space used: 24294472052
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146571.1


