

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
(338 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
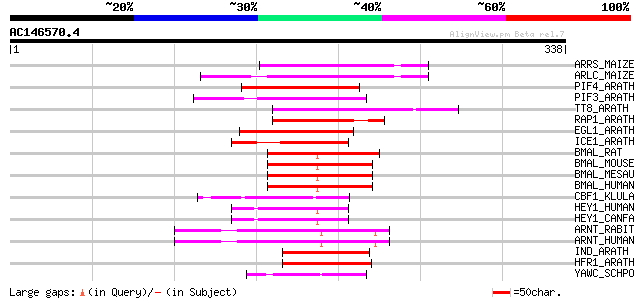
Score E
Sequences producing significant alignments: (bits) Value
ARRS_MAIZE (P13027) Anthocyanin regulatory R-S protein 60 8e-09
ARLC_MAIZE (P13526) Anthocyanin regulatory Lc protein 59 2e-08
PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4 (Basic heli... 57 9e-08
PIF3_ARATH (O80536) Phytochrome-interacting factor 3 (Phytochrom... 56 1e-07
TT8_ARATH (Q9FT81) TRANSPARENT TESTA 8 protein (Basic helix-loop... 54 7e-07
RAP1_ARATH (Q39204) Transcription factor AtMYC2 (R-homologous Ar... 54 7e-07
EGL1_ARATH (Q9CAD0) Transcription factor EGL1 (ENHANCER OF GLABR... 50 1e-05
ICE1_ARATH (Q9LSE2) Transcription factor ICE1 (Inducer of CBF ex... 49 1e-05
BMAL_RAT (Q9EPW1) Aryl hydrocarbon receptor nuclear translocator... 49 2e-05
BMAL_MOUSE (Q9WTL8) Aryl hydrocarbon receptor nuclear translocat... 48 3e-05
BMAL_MESAU (O88529) Aryl hydrocarbon receptor nuclear translocat... 48 3e-05
BMAL_HUMAN (O00327) Aryl hydrocarbon receptor nuclear translocat... 48 3e-05
CBF1_KLULA (P49379) Centromere-binding protein 1 (CBP-1) (Centro... 45 2e-04
HEY1_HUMAN (Q9Y5J3) Hairy/enhancer-of-split related with YRPW mo... 45 3e-04
HEY1_CANFA (Q9TSZ2) Hairy/enhancer-of-split related with YRPW mo... 45 3e-04
ARNT_RABIT (O02748) Aryl hydrocarbon receptor nuclear translocat... 45 3e-04
ARNT_HUMAN (P27540) Aryl hydrocarbon receptor nuclear translocat... 45 3e-04
IND_ARATH (O81313) INDEHISCENT protein 45 3e-04
HFR1_ARATH (Q9FE22) Long hypocotyl in far-red 1 (bHLH-like prote... 44 4e-04
YAWC_SCHPO (Q10186) Hypothetical protein C3F10.12c in chromosome I 44 8e-04
>ARRS_MAIZE (P13027) Anthocyanin regulatory R-S protein
Length = 612
Score = 60.1 bits (144), Expect = 8e-09
Identities = 33/103 (32%), Positives = 57/103 (55%), Gaps = 4/103 (3%)
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSL 212
A+ ++A+K+H +ER+RRE++N L+SLLPS + +KAS+LAE I ++KEL+R+
Sbjct: 410 AQEMSATKNHVMSERKRREKLNEMFLVLKSLLPSIHRVNKASILAETIAYLKELQRRVQE 469
Query: 213 IAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDR 255
+ + + E T S GN ++ +C +
Sbjct: 470 LESSREPASRPSETTTRLITRP----SRGNNESVRKEVCAGSK 508
>ARLC_MAIZE (P13526) Anthocyanin regulatory Lc protein
Length = 610
Score = 58.5 bits (140), Expect = 2e-08
Identities = 39/139 (28%), Positives = 64/139 (45%), Gaps = 13/139 (9%)
Query: 117 EHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNH 176
E R++ + G GG G +MS +K+H +ER+RRE++N
Sbjct: 381 EPQRLLKKVVAGGGAWESCGGATGAAQEMSG---------TGTKNHVMSERKRREKLNEM 431
Query: 177 LAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELTVDAAADDED 236
L+SLLPS + +KAS+LAE I ++KEL+R+ + + + E T
Sbjct: 432 FLVLKSLLPSIHRVNKASILAETIAYLKELQRRVQELESSREPASRPSETTTRLITRP-- 489
Query: 237 YGSNGNKFIIKASLCCDDR 255
S GN ++ +C +
Sbjct: 490 --SRGNNESVRKEVCAGSK 506
>PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4 (Basic
helix-loop-helix protein 9) (bHLH9) (AtbHLH009) (Short
under red-light 2)
Length = 430
Score = 56.6 bits (135), Expect = 9e-08
Identities = 28/72 (38%), Positives = 45/72 (61%)
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G S Q + A++ H+ +ERRRR+RIN + L+ L+P +KTDKAS+L E I
Sbjct: 242 IGNKSNQRSGSNRRSRAAEVHNLSERRRRDRINERMKALQELIPHCSKTDKASILDEAID 301
Query: 202 HVKELKRQTSLI 213
++K L+ Q ++
Sbjct: 302 YLKSLQLQLQVM 313
>PIF3_ARATH (O80536) Phytochrome-interacting factor 3
(Phytochrome-associated protein 3) (Basic
helix-loop-helix protein 8) (bHLH8) (AtbHLH008)
Length = 524
Score = 55.8 bits (133), Expect = 1e-07
Identities = 31/105 (29%), Positives = 57/105 (53%), Gaps = 7/105 (6%)
Query: 113 HAGSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRER 172
H+ + + S+ ++ SG G G + +K +++ H+ +ERRRR+R
Sbjct: 306 HSNIQDIDCHSEDVEEESGDGRKEAGPSRTG-------LGSKRSRSAEVHNLSERRRRDR 358
Query: 173 INNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETS 217
IN + L+ L+P+ K DKAS+L E I+++K L+ Q +++ S
Sbjct: 359 INEKMRALQELIPNCNKVDKASMLDEAIEYLKSLQLQVQIMSMAS 403
>TT8_ARATH (Q9FT81) TRANSPARENT TESTA 8 protein (Basic
helix-loop-helix protein 42) (bHLH42) (AtbHLH042)
Length = 518
Score = 53.5 bits (127), Expect = 7e-07
Identities = 34/113 (30%), Positives = 55/113 (48%), Gaps = 1/113 (0%)
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP 220
SH AERRRRE++N LRS++P TK DK S+L + I +V L+++ + T
Sbjct: 363 SHVVAERRRREKLNEKFITLRSMVPFVTKMDKVSILGDTIAYVNHLRKRVHELENTHHEQ 422
Query: 221 TECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRT 273
T +E S ++ + C+ R LL ++++ L L + T
Sbjct: 423 QHKRTRTCKRKTSEEVEVSIIENDVL-LEMRCEYRDGLLLDILQVLHELGIET 474
>RAP1_ARATH (Q39204) Transcription factor AtMYC2 (R-homologous
Arabidopsis protein-1) (RAP-1) (Basic helix-loop-helix
protein 6) (bHLH6) (AtbHLH006) (rd22BP1)
Length = 623
Score = 53.5 bits (127), Expect = 7e-07
Identities = 27/68 (39%), Positives = 43/68 (62%), Gaps = 8/68 (11%)
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP 220
+H EAER+RRE++N LR+++P+ +K DKASLL + I ++ ELK + V
Sbjct: 452 NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAIAYINELKSKV--------VK 503
Query: 221 TECDELTV 228
TE ++L +
Sbjct: 504 TESEKLQI 511
>EGL1_ARATH (Q9CAD0) Transcription factor EGL1 (ENHANCER OF GLABRA3)
(Basic helix-loop-helix protein 2) (bHLH2) (AtbHLH002)
(AtMyc-146)
Length = 596
Score = 49.7 bits (117), Expect = 1e-05
Identities = 22/69 (31%), Positives = 46/69 (65%)
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
EV M+ +E + + +H+ +E++RRE++N LRS++PS +K DK S+L + I
Sbjct: 385 EVPLMNKKEELLPDTPEETGNHALSEKKRREKLNERFMTLRSIIPSISKIDKVSILDDTI 444
Query: 201 QHVKELKRQ 209
+++++L+++
Sbjct: 445 EYLQDLQKR 453
>ICE1_ARATH (Q9LSE2) Transcription factor ICE1 (Inducer of CBF
expression 1) (Basic helix-loop-helix protein 116)
(bHLH116) (AtbHLH116)
Length = 494
Score = 49.3 bits (116), Expect = 1e-05
Identities = 27/71 (38%), Positives = 43/71 (60%), Gaps = 13/71 (18%)
Query: 136 GGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASL 195
GG +G+ M A+ +M AERRRR+++N+ L LRS++P +K D+AS+
Sbjct: 295 GGGKGKKKGMPAKNLM-------------AERRRRKKLNDRLYMLRSVVPKISKMDRASI 341
Query: 196 LAEVIQHVKEL 206
L + I ++KEL
Sbjct: 342 LGDAIDYLKEL 352
>BMAL_RAT (Q9EPW1) Aryl hydrocarbon receptor nuclear
translocator-like protein 1 (Brain and muscle ARNT-like
1) (Tic)
Length = 626
Score = 48.5 bits (114), Expect = 2e-05
Identities = 24/72 (33%), Positives = 45/72 (62%), Gaps = 4/72 (5%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPS----TTKTDKASLLAEVIQHVKELKRQTSLI 213
A ++HS+ E+RRR+++N+ + +L SL+P+ + K DK ++L +QH+K L+ T+
Sbjct: 73 AREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPY 132
Query: 214 AETSPVPTECDE 225
E + PT +
Sbjct: 133 TEANYKPTSLSD 144
>BMAL_MOUSE (Q9WTL8) Aryl hydrocarbon receptor nuclear
translocator-like protein 1 (Brain and muscle ARNT-like
1) (Arnt3)
Length = 632
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/68 (35%), Positives = 44/68 (64%), Gaps = 4/68 (5%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPS----TTKTDKASLLAEVIQHVKELKRQTSLI 213
A ++HS+ E+RRR+++N+ + +L SL+P+ + K DK ++L +QH+K L+ T+
Sbjct: 80 AREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPY 139
Query: 214 AETSPVPT 221
E + PT
Sbjct: 140 TEANYKPT 147
>BMAL_MESAU (O88529) Aryl hydrocarbon receptor nuclear
translocator-like protein 1 (Brain and muscle ARNT-like
1)
Length = 626
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/68 (35%), Positives = 44/68 (64%), Gaps = 4/68 (5%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPS----TTKTDKASLLAEVIQHVKELKRQTSLI 213
A ++HS+ E+RRR+++N+ + +L SL+P+ + K DK ++L +QH+K L+ T+
Sbjct: 73 AREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPY 132
Query: 214 AETSPVPT 221
E + PT
Sbjct: 133 TEANYKPT 140
>BMAL_HUMAN (O00327) Aryl hydrocarbon receptor nuclear
translocator-like protein 1 (Brain and muscle ARNT-like
1) (Member of PAS protein 3) (Basic-helix-loop-helix-PAS
orphan MOP3) (bHLH-PAS protein JAP3)
Length = 626
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/68 (35%), Positives = 44/68 (64%), Gaps = 4/68 (5%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPS----TTKTDKASLLAEVIQHVKELKRQTSLI 213
A ++HS+ E+RRR+++N+ + +L SL+P+ + K DK ++L +QH+K L+ T+
Sbjct: 73 AREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPY 132
Query: 214 AETSPVPT 221
E + PT
Sbjct: 133 TEANYKPT 140
>CBF1_KLULA (P49379) Centromere-binding protein 1 (CBP-1)
(Centromere-binding factor 1) (Centromere promoter
factor 1)
Length = 359
Score = 45.4 bits (106), Expect = 2e-04
Identities = 30/93 (32%), Positives = 48/93 (51%), Gaps = 7/93 (7%)
Query: 115 GSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERIN 174
G+EH D G+G+G +G K + A +SH E ERRRR+ IN
Sbjct: 213 GTEH----DDEENGGAGAGGAAPRRGR--KPGTETGSTAWKQQRKESHKEVERRRRQNIN 266
Query: 175 NHLAKLRSLLPSTTKTDKASLLAEVIQHVKELK 207
+ KL LLP +T KA++L+ ++++++K
Sbjct: 267 TAIEKLSDLLP-VKETSKAAILSRAAEYIQKMK 298
>HEY1_HUMAN (Q9Y5J3) Hairy/enhancer-of-split related with YRPW motif
1 (Hairy and enhancer of split related-1) (HESR-1)
(Cardiovascular helix-loop-helix factor 2) (HES-related
repressor protein 2 HERP2)
Length = 304
Score = 45.1 bits (105), Expect = 3e-04
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 8/77 (10%)
Query: 136 GGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPS------TTK 189
G +G MS ++ LA + E+RRR+RINN L++LR L+PS + K
Sbjct: 30 GNLSSALGSMSPTT--SSQILARKRRRGIIEKRRRDRINNSLSELRRLVPSAFEKQGSAK 87
Query: 190 TDKASLLAEVIQHVKEL 206
+KA +L + H+K L
Sbjct: 88 LEKAEILQMTVDHLKML 104
>HEY1_CANFA (Q9TSZ2) Hairy/enhancer-of-split related with YRPW motif
1 (Hairy and enhancer of split related-1) (HESR-1)
Length = 304
Score = 45.1 bits (105), Expect = 3e-04
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 8/77 (10%)
Query: 136 GGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPS------TTK 189
G +G MS ++ LA + E+RRR+RINN L++LR L+PS + K
Sbjct: 30 GNLSSALGSMSPTT--SSQILARKRRRGIIEKRRRDRINNSLSELRRLVPSAFEKQGSAK 87
Query: 190 TDKASLLAEVIQHVKEL 206
+KA +L + H+K L
Sbjct: 88 LEKAEILQMTVDHLKML 104
>ARNT_RABIT (O02748) Aryl hydrocarbon receptor nuclear translocator
(ARNT protein) (Dioxin receptor, nuclear translocator)
(Hypoxia-inducible factor 1 beta) (HIF-1 beta)
Length = 790
Score = 45.1 bits (105), Expect = 3e-04
Identities = 34/140 (24%), Positives = 63/140 (44%), Gaps = 18/140 (12%)
Query: 101 RRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASK 160
+R P L F D S+ LR D + + + + ++ K A +
Sbjct: 42 KRRPGLDFDDDGEGNSKFLRCDEDQMSND---------KERFARSDDEQSSADKERLARE 92
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTT----KTDKASLLAEVIQHVKELKRQTSLIAET 216
+HSE ERRRR ++ ++ +L ++P+ + K DK ++L + H+K L+ + +
Sbjct: 93 NHSEIERRRRNKMTAYITELSDIVPTCSALARKPDKLTILRMAVSHMKSLRGTGNTSTDG 152
Query: 217 SPVPT-----ECDELTVDAA 231
S P+ E L ++AA
Sbjct: 153 SYKPSFLTDQELKHLILEAA 172
>ARNT_HUMAN (P27540) Aryl hydrocarbon receptor nuclear translocator
(ARNT protein) (Dioxin receptor, nuclear translocator)
(Hypoxia-inducible factor 1 beta) (HIF-1 beta)
Length = 789
Score = 45.1 bits (105), Expect = 3e-04
Identities = 34/140 (24%), Positives = 63/140 (44%), Gaps = 18/140 (12%)
Query: 101 RRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASK 160
+R P L F D S+ LR D + + + + ++ K A +
Sbjct: 42 KRRPGLDFDDDGEGNSKFLRCDDDQMSND---------KERFARSDDEQSSADKERLARE 92
Query: 161 SHSEAERRRRERINNHLAKLRSLLPSTT----KTDKASLLAEVIQHVKELKRQTSLIAET 216
+HSE ERRRR ++ ++ +L ++P+ + K DK ++L + H+K L+ + +
Sbjct: 93 NHSEIERRRRNKMTAYITELSDMVPTCSALARKPDKLTILRMAVSHMKSLRGTGNTSTDG 152
Query: 217 SPVPT-----ECDELTVDAA 231
S P+ E L ++AA
Sbjct: 153 SYKPSFLTDQELKHLILEAA 172
>IND_ARATH (O81313) INDEHISCENT protein
Length = 169
Score = 44.7 bits (104), Expect = 3e-04
Identities = 22/53 (41%), Positives = 33/53 (61%)
Query: 167 RRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPV 219
RRRRERI+ + L+ ++P K D AS+L E I++ K LKRQ ++ S +
Sbjct: 99 RRRRERISEKIRILKRIVPGGAKMDTASMLDEAIRYTKFLKRQVRILQPHSQI 151
>HFR1_ARATH (Q9FE22) Long hypocotyl in far-red 1 (bHLH-like protein
HFR1) (Reduced phytochrome signaling) (Basic
helix-loop-helix FBI1 protein) (Basic helix-loop-helix
protein 26) (bHLH26) (AtbHLH026) (Reduced sensitivity to
far-red light)
Length = 292
Score = 44.3 bits (103), Expect = 4e-04
Identities = 20/54 (37%), Positives = 36/54 (66%)
Query: 167 RRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVP 220
RRR E+++N + KL+ L+P+ KTDK S+L + I+++K L+ Q +++ P
Sbjct: 144 RRRDEKMSNKMRKLQQLVPNCHKTDKVSVLDKTIEYMKNLQLQLQMMSTVGVNP 197
>YAWC_SCHPO (Q10186) Hypothetical protein C3F10.12c in chromosome I
Length = 201
Score = 43.5 bits (101), Expect = 8e-04
Identities = 25/73 (34%), Positives = 41/73 (55%), Gaps = 5/73 (6%)
Query: 145 MSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVK 204
+ ++E +AK L SH E ERRRRE I+ + +L +++P K +K S+L Q+++
Sbjct: 80 VGSEEWYKAKRL----SHKEVERRRREAISEGIKELANIVPGCEK-NKGSILQRTAQYIR 134
Query: 205 ELKRQTSLIAETS 217
LK + E S
Sbjct: 135 SLKEMEEMCREKS 147
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,066,911
Number of Sequences: 164201
Number of extensions: 1594891
Number of successful extensions: 7078
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 6643
Number of HSP's gapped (non-prelim): 382
length of query: 338
length of database: 59,974,054
effective HSP length: 111
effective length of query: 227
effective length of database: 41,747,743
effective search space: 9476737661
effective search space used: 9476737661
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146570.4


