

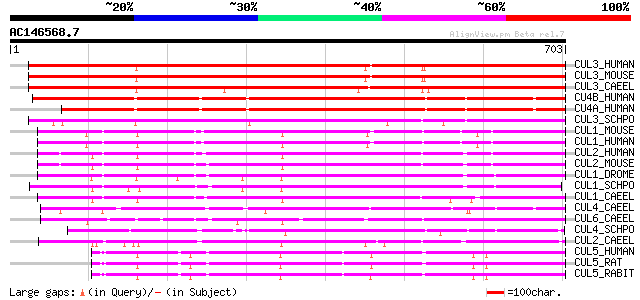
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.7 - phase: 0
(703 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CUL3_HUMAN (Q13618) Cullin homolog 3 (CUL-3) 694 0.0
CUL3_MOUSE (Q9JLV5) Cullin homolog 3 (CUL-3) 693 0.0
CUL3_CAEEL (Q17391) Cullin 3 531 e-150
CU4B_HUMAN (Q13620) Cullin homolog 4B (CUL-4B) 491 e-138
CU4A_HUMAN (Q13619) Cullin homolog 4A (CUL-4A) 484 e-136
CUL3_SCHPO (Q09760) Cullin 3 homolog (Cul-3) 444 e-124
CUL1_MOUSE (Q9WTX6) Cullin homolog 1 (CUL-1) 374 e-103
CUL1_HUMAN (Q13616) Cullin homolog 1 (CUL-1) 374 e-103
CUL2_HUMAN (Q13617) Cullin homolog 2 (CUL-2) 345 2e-94
CUL2_MOUSE (Q9D4H8) Cullin homolog 2 (CUL-2) 340 8e-93
CUL1_DROME (Q24311) Cullin homolog 1 (Lin-19 homolog protein) 331 3e-90
CUL1_SCHPO (O13790) Cullin 1 homolog (Cul-1) (Cell division cont... 320 1e-86
CUL1_CAEEL (Q17389) Cullin 1 (Abnormal cell lineage 19 protein) 316 2e-85
CUL4_CAEEL (Q17392) Cullin 4 287 6e-77
CUL6_CAEEL (Q21346) Cullin 6 283 1e-75
CUL4_SCHPO (O14122) Cullin 4 homolog (Cul-4) 283 1e-75
CUL2_CAEEL (Q17390) Cullin 2 275 4e-73
CUL5_HUMAN (Q93034) Cullin homolog 5 (CUL-5) (Vasopressin-activa... 229 2e-59
CUL5_RAT (Q9JJ31) Cullin homolog 5 (CUL-5) (Vasopressin-activate... 225 4e-58
CUL5_RABIT (Q29425) Cullin homolog 5 (CUL-5) (Vasopressin-activa... 224 6e-58
>CUL3_HUMAN (Q13618) Cullin homolog 3 (CUL-3)
Length = 768
Score = 694 bits (1791), Expect = 0.0
Identities = 371/712 (52%), Positives = 502/712 (70%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLG 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLL 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 536
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 537 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 596
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 597 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 656
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 657 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 716
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 717 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>CUL3_MOUSE (Q9JLV5) Cullin homolog 3 (CUL-3)
Length = 768
Score = 693 bits (1789), Expect = 0.0
Identities = 370/712 (51%), Positives = 501/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSCYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 536
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 537 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 596
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 597 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 656
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 657 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 716
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 717 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>CUL3_CAEEL (Q17391) Cullin 3
Length = 777
Score = 531 bits (1368), Expect = e-150
Identities = 299/724 (41%), Positives = 448/724 (61%), Gaps = 45/724 (6%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIA-KSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G+RLY+GL + H+ + + IE+ GSFLE + W DH A+
Sbjct: 55 YRNAYTMVLHKHGERLYNGLKDVIQDHMASVRIRIIESMNSGSFLETVAESWADHTVAMV 114
Query: 83 MIRDILMYMDRTFIPSAKKT-PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGE 141
MIRDILMYMDR ++ PV+ LGL+ +R ++ N I R+ + LLEL++ +R
Sbjct: 115 MIRDILMYMDRIYVAQNNHVLPVYNLGLDAYRTEILRQNGIGDRIRDALLELIKLDRKSN 174
Query: 142 VIDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECC-DCGDYLK 197
I+ ++N ML+ LG VY +FE L+ ++++Y+ + ++ D YL
Sbjct: 175 QINWHGIKNACDMLISLGIDSRTVYEDEFERPLLKETSDYYRDVCKNWLSGDNDACFYLA 234
Query: 198 KAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYED 257
+ E +++E R Y+D TE KI +V++ M+ H+ +++M+N G+ ML K ED
Sbjct: 235 QVEIAMHDEASRASRYLDKMTEAKILQVMDDVMVAEHIQTIVYMQNGGVKFMLEHKKIED 294
Query: 258 LGRMYNLFRRVAD-------GLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDE 310
L R++ +F+R+ D GL + + ++ ++ E+G +V + + LK+PV FV LL
Sbjct: 295 LTRIFRIFKRIGDSVTVPGGGLKALLKAVSEYLNETGSNIVKNEDLLKNPVNFVNELLQL 354
Query: 311 KDKYDKIINQAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDV 370
KD + ++ AF +D+ F+N FE F+N N +SPEF++L++DD LR GLK V++ ++
Sbjct: 355 KDYFSSLLTTAFADDRDFKNRFQHDFETFLNSNRQSPEFVALYMDDMLRSGLKCVSDAEM 414
Query: 371 EVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTS 430
+ LD VM+LFRYLQEKDVFEKY+KQ+LAKRLL K+ SDD E++L+ KLKTECG QFT
Sbjct: 415 DNKLDNVMILFRYLQEKDVFEKYFKQYLAKRLLLDKSCSDDVEKALLAKLKTECGCQFTQ 474
Query: 431 KLEGMFTDMK---TSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEIS 487
KLE MF D + T + + + + P ++++VLT G WPT LP E+S
Sbjct: 475 KLENMFRDKELWLTLATSFRDWREAQP-TKMSIDISLRVLTAGVWPTVQCNPVVLPQELS 533
Query: 488 ALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATF--------------------GKGQKH 527
E F YY HTGR+L+ T +G AD+KATF G+ K
Sbjct: 534 VAYEMFTQYYTEKHTGRKLTINTLLGNADVKATFYPPPKASMSNEENGPGPSSSGESMKE 593
Query: 528 E------LNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKG--RNV 579
L V+T+QM +L+ FN+ +++S +++ +IP +LKR LQSLAL K R +
Sbjct: 594 RKPEHKILQVNTHQMIILLQFNHHNRISCQQLMDELKIPERELKRNLQSLALGKASQRIL 653
Query: 580 LRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIE 639
+RK + D F+VND F SKL +VK+ V + ESEPE +ETRQ+VE+DRK ++E
Sbjct: 654 VRKNKGKDAIDMSDEFAVNDNFQSKLTRVKVQMVTGKVESEPEIRETRQKVEDDRKLEVE 713
Query: 640 AAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMY 699
AAIVRIMK+R+ L+HNNL+AEVT+QL+ RF+ +P +K+RIE+LIER++L RD++D + Y
Sbjct: 714 AAIVRIMKARKKLNHNNLVAEVTQQLRHRFMPSPIIIKQRIETLIEREYLARDEHDHRAY 773
Query: 700 RYLA 703
+Y+A
Sbjct: 774 QYIA 777
>CU4B_HUMAN (Q13620) Cullin homolog 4B (CUL-4B)
Length = 895
Score = 491 bits (1265), Expect = e-138
Identities = 277/681 (40%), Positives = 416/681 (60%), Gaps = 19/681 (2%)
Query: 29 NMVLHKFGDRLYSGLVATMTAHLKEIAKSI--EAAQGGSFLEELNRKWNDHNKALQMIRD 86
N+ +K LY L H+K ++ FL++++R W +H + + MIR
Sbjct: 228 NLCSYKISANLYKQLRQICEDHIKAQIHQFREDSLDSVLFLKKIDRCWQNHCRQMIMIRS 287
Query: 87 ILMYMDRTFIPSAKKTP-VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDR 145
I +++DRT++ P + ++GL L+R +I +++ + ++ +L L++ ER GE IDR
Sbjct: 288 IFLFLDRTYVLQNSMLPSIWDMGLELFRAHIISDQKVQNKTIDGILLLIERERNGEAIDR 347
Query: 146 GIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNE 205
++R++ ML DL +Y FE FL+ + Y E Q+ ++ + +YL +RL E
Sbjct: 348 SLLRSLLSMLSDL--QIYQDSFEQRFLEETNRLYAAEGQKLMQEREVPEYLHHVNKRLEE 405
Query: 206 EMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLF 265
E DR+ Y+D T+K + VE Q++ H+ ++ GL N+L +++ +DL +Y LF
Sbjct: 406 EADRLITYLDQTTQKSLIATVEKQLLGEHLTAILQ---KGLNNLLDENRIQDLSLLYQLF 462
Query: 266 RRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNND 325
RV G+ + + +I+ G +V +PE+ K VQ LLD KDK D II+ F +
Sbjct: 463 SRVRGGVQVLLQQWIEYIKAFGSTIVINPEKDKT---MVQELLDFKDKVDHIIDICFLKN 519
Query: 326 KSFQNALNSSFEYFINLNPRSP-EFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYL 384
+ F NA+ +FE FIN P P E I+ +VD KLR G K ++++E LDK+M++FR++
Sbjct: 520 EKFINAMKEAFETFINKRPNKPAELIAKYVDSKLRAGNKEATDEELEKMLDKIMIIFRFI 579
Query: 385 QEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQD 444
KDVFE +YK+ LAKRLL GK+ S DAE+S++ KLK ECG FTSKLEGMF DM+ S+D
Sbjct: 580 YGKDVFEAFYKKDLAKRLLVGKSASVDAEKSMLSKLKHECGAAFTSKLEGMFKDMELSKD 639
Query: 445 TMQGF--YASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHT 502
M F Y + ++ LTV +LT G WPT + +LP E+ L E F+++YLG H+
Sbjct: 640 IMIQFKQYMQNQNVPGNIELTVNILTMGYWPTYVPMEVHLPPEMVKLQEIFKTFYLGKHS 699
Query: 503 GRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAP 562
GR+L WQ+ +G LKA F +G+K EL VS +Q VL++FN ++ S +EI+QAT I
Sbjct: 700 GRKLQWQSTLGHCVLKAEFKEGKK-ELQVSLFQTLVLLMFNEGEEFSLEEIKQATGIEDG 758
Query: 563 DLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPE 622
+L+R LQSLA K R VL K P KD+ + D F ND F KL+++KI + KE+ E
Sbjct: 759 ELRRTLQSLACGKAR-VLAKNPKGKDIEDGDKFICNDDFKHKLFRIKINQ-IQMKETVEE 816
Query: 623 KQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIES 682
+ T +RV +DR+ QI+AAIVRIMK R+ L HN L++EV QL+F P ++KKRIES
Sbjct: 817 QASTTERVFQDRQYQIDAAIVRIMKMRKTLSHNLLVSEVYN--QLKFPVKPADLKKRIES 874
Query: 683 LIERDFLERDDNDRKMYRYLA 703
LI+RD++ERD + Y Y+A
Sbjct: 875 LIDRDYMERDKENPNQYNYIA 895
>CU4A_HUMAN (Q13619) Cullin homolog 4A (CUL-4A)
Length = 659
Score = 484 bits (1247), Expect = e-136
Identities = 275/642 (42%), Positives = 403/642 (61%), Gaps = 17/642 (2%)
Query: 66 FLEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTP-VHELGLNLWRESVIYSNQIRT 124
FL+++N W DH + + MIR I +++DRT++ P + ++GL L+R +I +++
Sbjct: 31 FLKKINTCWQDHCRQMIMIRSIFLFLDRTYVLQNSTLPSIWDMGLELFRTHIISDKMVQS 90
Query: 125 RLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQ 184
+ ++ +L L++ ER+GE +DR ++R++ ML DL VY FE FL+ + Y E Q
Sbjct: 91 KTIDGILLLIERERSGEAVDRSLLRSLLGMLSDL--QVYKDSFELKFLEETNCLYAAEGQ 148
Query: 185 RFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENS 244
R ++ + +YL +RL EE DRV Y+D T+K + VE Q++ H+ ++
Sbjct: 149 RLMQEREVPEYLNHVSKRLEEEGDRVITYLDHSTQKPLIACVEKQLLGEHLTAILQ---K 205
Query: 245 GLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFV 304
GL ++L +++ DL +MY LF RV G + + + +I+ G +V +PE+ KD V
Sbjct: 206 GLDHLLDENRVPDLAQMYQLFSRVRGGQQALLQHWSEYIKTFGTAIVINPEKDKD---MV 262
Query: 305 QRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLNPRSP-EFISLFVDDKLRKGLK 363
Q LLD KDK D +I F ++ F N + SFE FIN P P E I+ VD KLR G K
Sbjct: 263 QDLLDFKDKVDHVIEVCFQKNERFVNLMKESFETFINKRPNKPAELIAKHVDSKLRAGNK 322
Query: 364 GVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTE 423
++++E TLDK+M+LFR++ KDVFE +YK+ LAKRLL GK+ S DAE+S++ KLK E
Sbjct: 323 EATDEELERTLDKIMILFRFIHGKDVFEAFYKKDLAKRLLVGKSASVDAEKSMLSKLKHE 382
Query: 424 CGYQFTSKLEGMFTDMKTSQDTMQGFYASHPDLGD-GP-TLTVQVLTTGSWPTQSSITCN 481
CG FTSKLEGMF DM+ S+D M F + D GP LTV +LT G WPT + + +
Sbjct: 383 CGAAFTSKLEGMFKDMELSKDIMVHFKQHMQNQSDSGPIDLTVNILTMGYWPTYTPMEVH 442
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLML 541
L E+ L E F+++YLG H+GR+L WQT +G A LKA F +G+K E VS +Q VL++
Sbjct: 443 LTPEMIKLQEVFKAFYLGKHSGRKLQWQTTLGHAVLKAEFKEGKK-EFQVSLFQTLVLLM 501
Query: 542 FNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKF 601
FN D S++EI+ AT I +L+R LQSLA K R VL K P K+V + D F N +F
Sbjct: 502 FNEGDGFSFEEIKMATGIEDSELRRTLQSLACGKAR-VLIKSPKGKEVEDGDKFIFNGEF 560
Query: 602 SSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEV 661
KL+++KI + KE+ E+ T +RV +DR+ QI+AAIVRIMK R+ L HN L++E+
Sbjct: 561 KHKLFRIKINQ-IQMKETVEEQVSTTERVFQDRQYQIDAAIVRIMKMRKTLGHNLLVSEL 619
Query: 662 TKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
QL+F P ++KKRIESLI+RD++ERD ++ Y Y+A
Sbjct: 620 YN--QLKFPVKPGDLKKRIESLIDRDYMERDKDNPNQYHYVA 659
>CUL3_SCHPO (Q09760) Cullin 3 homolog (Cul-3)
Length = 785
Score = 444 bits (1143), Expect = e-124
Identities = 266/738 (36%), Positives = 419/738 (56%), Gaps = 60/738 (8%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKE-----IAKSIEAAQGGS------------- 65
+RNAY +VLHK+G++LY+ + + + LKE I K+ +A+ G+
Sbjct: 50 YRNAYILVLHKYGEKLYNHVQDVIRSRLKEETVPAIYKNYDASLLGNALLDIRKNDSYST 109
Query: 66 -----------FLEELNRKWNDHNKALQMIRDILMYMDRTFIPSAKKTPVHELGLNLWRE 114
FL L W DH ++QMI +L Y+D+ + SA K PV+E G+ ++RE
Sbjct: 110 SWSRSLEAAHRFLSSLVNSWKDHIVSMQMISSVLKYLDKVYSKSADKVPVNENGIYIFRE 169
Query: 115 SVIYSN-QIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDL-----GPAVYGQDFE 168
V+ ++ +I + + T+L LV ER G I+R ++ + ML L +Y F
Sbjct: 170 VVLLNSFEIGEKCVETILILVYLERKGNTINRPLINDCLDMLNSLPSENKKETLYDVLFA 229
Query: 169 AHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVET 228
FL + FY++ES I +YLKKAE+R EE +R +Y+ + + VVE
Sbjct: 230 PKFLSYTRNFYEIESSTVIGVFGVVEYLKKAEKRFEEEKERSKNYLFTKIASPLLSVVED 289
Query: 229 QMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGK 288
+++ H+ L+ +++G +M+ +E L +Y F RV G+ +++ + ++ GK
Sbjct: 290 ELLSKHLDDLLENQSTGFFSMIDSSNFEGLQLVYESFSRVELGVKSLKKYLAKYVAHHGK 349
Query: 289 QL-VTDPERLKDPVE-------------FVQRLLDEKDKYDKIINQAFNNDKSFQNALNS 334
+ T + L+ + +VQ++L D+ + II+ + D+S N+L+
Sbjct: 350 LINETTSQALEGKMAVGRLSSNATMATLWVQKVLALWDRLNTIISTTMDADRSILNSLSD 409
Query: 335 SFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYY 394
+F F++ R+PE+ISLF+DD L+K + E +E TL + LFR++ EKDVFEKYY
Sbjct: 410 AFVTFVDGYTRAPEYISLFIDDNLKKDARKAIEGSIEATLQNSVTLFRFISEKDVFEKYY 469
Query: 395 KQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHP 454
K HLAKRLL+ +++S DAE +I +LK E G FT KLEGMF DM SQ+ +Q + +
Sbjct: 470 KTHLAKRLLNNRSISSDAELGMISRLKQEAGNVFTQKLEGMFNDMNLSQELLQEYKHNSA 529
Query: 455 DLGDGPT--LTVQVLTTGSWPTQSS---ITCNLPVEISALCEKFRSYYLGTHTGRRLSWQ 509
P L V +L + WP S I CN P + A ++F +YL HTGR+L W
Sbjct: 530 LQSAKPALDLNVSILASTFWPIDLSPHKIKCNFPKVLLAQIDQFTDFYLSKHTGRKLLWY 589
Query: 510 TNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADK---LSYKEIEQATEIPAPDLKR 566
+MG AD++ F K +K++LNVST +L+LF + + L ++EI + T I DLKR
Sbjct: 590 PSMGSADVRVNF-KDRKYDLNVSTIASVILLLFQDLKENQCLIFEEILEKTNIEVGDLKR 648
Query: 567 CLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQK-ESEPEKQE 625
LQSLA K + +L K+P ++V D F N+ F S L ++KI TV + E + E++
Sbjct: 649 NLQSLACAKYK-ILLKDPKGREVNAGDKFYFNENFVSNLARIKISTVAQTRVEDDSERKR 707
Query: 626 TRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIE 685
T ++V+E RK Q +A IVR+MK R++ +HN L+AEVT+QL RF +P +K+RIE+LIE
Sbjct: 708 TLEKVDESRKHQADACIVRVMKDRKVCEHNQLMAEVTRQLNPRFHPSPMMIKRRIEALIE 767
Query: 686 RDFLERDDNDRKMYRYLA 703
R++L+R ++ ++Y YLA
Sbjct: 768 REYLQRQADNGRIYEYLA 785
>CUL1_MOUSE (Q9WTX6) Cullin homolog 1 (CUL-1)
Length = 776
Score = 374 bits (961), Expect = e-103
Identities = 241/705 (34%), Positives = 382/705 (54%), Gaps = 50/705 (7%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTF 95
G LY L + +L + K E S L+ ++W D+ + +++ I Y++R +
Sbjct: 85 GLELYKRLKEFLKNYLTNLLKDGEDLMDESVLKFYTQQWEDYRFSSKVLNGICAYLNRHW 144
Query: 96 I----PSAKKT--PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMR 149
+ +K ++ L L WR+ + + ++ N +L+L++ ER GE I+ ++
Sbjct: 145 VRRECDEGRKGIYEIYSLALVTWRDCLF--RPLNKQVTNAVLKLIEKERNGETINTRLIS 202
Query: 150 NITKMLMDLGP------------AVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLK 197
+ + ++LG VY + FE+ FL + FY ES F++ +Y+K
Sbjct: 203 GVVQSYVELGLNEDDAFAKGPTLTVYKESFESQFLADTERFYTRESTEFLQQNPVTEYMK 262
Query: 198 KAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYED 257
KAE RL EE RV Y+ T+ ++ + E +IE H L + H E N+L DK ED
Sbjct: 263 KAEARLLEEQRRVQVYLHESTQDELARKCEQVLIEKH-LEIFHTE---FQNLLDADKNED 318
Query: 258 LGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTD--PERLKDPVEFVQRLLDEKDKYD 315
LGRMYNL R+ DGL ++++++ HI G + L DP +VQ +LD KY+
Sbjct: 319 LGRMYNLVSRIQDGLGELKKLLETHIHNQGLAAIEKCGEAALNDPKMYVQTVLDVHKKYN 378
Query: 316 KIINQAFNNDKSFQNALNSSFEYFINLNP---------RSPEFISLFVDDKLRKGLKGVN 366
++ AFNND F AL+ + FIN N +SPE ++ + D L+K K
Sbjct: 379 ALVMSAFNNDAGFVAALDKACGRFINNNAVTKMAQSSSKSPELLARYCDSLLKKSSKNPE 438
Query: 367 EDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGY 426
E ++E TL++VM++F+Y+++KDVF+K+Y + LAKRL+ + SDDAE S+I KLK CG+
Sbjct: 439 EAELEDTLNQVMVVFKYIEDKDVFQKFYAKMLAKRLVHQNSASDDAEASMISKLKQACGF 498
Query: 427 QFTSKLEGMFTDMKTSQDTMQGFYA----SHP-DLGDGPTLTVQVLTTGSWPTQSSITCN 481
++TSKL+ MF D+ S+D + F S P DL ++QVL++GSWP Q S T
Sbjct: 499 EYTSKLQRMFQDIGVSKDLNEQFKKHLTNSEPLDL----DFSIQVLSSGSWPFQQSCTFA 554
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLML 541
LP E+ ++F ++Y H+GR+L+W + +L K ++ L ST+QM +L+
Sbjct: 555 LPSELERSYQRFTAFYASRHSGRKLTWLYQLSKGELVTNCFK-NRYTLQASTFQMAILLQ 613
Query: 542 FNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGE---DDAFSVN 598
+N D + +++ +T+I L + LQ L L VL E + D E D +
Sbjct: 614 YNTEDAYTVQQLTDSTQIKMDILAQVLQIL-LKSKLLVLEDENANVDEVELKPDTLIKLY 672
Query: 599 DKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLI 658
+ +K +V I V + E + E++ T + +EEDRK I+AAIVRIMK R++L H L+
Sbjct: 673 LGYKNKKLRVNI-NVPMKTEQKQEQETTHKNIEEDRKLLIQAAIVRIMKMRKVLKHQQLL 731
Query: 659 AEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
EV QL RF +KK I+ LIE+++LER D ++ Y YLA
Sbjct: 732 GEVLTQLSSRFKPRVPVIKKCIDILIEKEYLERVDGEKDTYSYLA 776
>CUL1_HUMAN (Q13616) Cullin homolog 1 (CUL-1)
Length = 776
Score = 374 bits (961), Expect = e-103
Identities = 241/705 (34%), Positives = 382/705 (54%), Gaps = 50/705 (7%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTF 95
G LY L + +L + K E S L+ ++W D+ + +++ I Y++R +
Sbjct: 85 GLELYKRLKEFLKNYLTNLLKDGEDLMDESVLKFYTQQWEDYRFSSKVLNGICAYLNRHW 144
Query: 96 I----PSAKKT--PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMR 149
+ +K ++ L L WR+ + + ++ N +L+L++ ER GE I+ ++
Sbjct: 145 VRRECDEGRKGIYEIYSLALVTWRDCLF--RPLNKQVTNAVLKLIEKERNGETINTRLIS 202
Query: 150 NITKMLMDLGP------------AVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLK 197
+ + ++LG VY + FE+ FL + FY ES F++ +Y+K
Sbjct: 203 GVVQSYVELGLNEDDAFAKGPTLTVYKESFESQFLADTERFYTRESTEFLQQNPVTEYMK 262
Query: 198 KAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYED 257
KAE RL EE RV Y+ T+ ++ + E +IE H L + H E N+L DK ED
Sbjct: 263 KAEARLLEEQRRVQVYLHESTQDELARKCEQVLIEKH-LEIFHTE---FQNLLDADKNED 318
Query: 258 LGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTD--PERLKDPVEFVQRLLDEKDKYD 315
LGRMYNL R+ DGL ++++++ HI G + L DP +VQ +LD KY+
Sbjct: 319 LGRMYNLVSRIQDGLGELKKLLETHIHNQGLAAIEKCGEAALNDPKMYVQTVLDVHKKYN 378
Query: 316 KIINQAFNNDKSFQNALNSSFEYFINLNP---------RSPEFISLFVDDKLRKGLKGVN 366
++ AFNND F AL+ + FIN N +SPE ++ + D L+K K
Sbjct: 379 ALVMSAFNNDAGFVAALDKACGRFINNNAVTKMAQSSSKSPELLARYCDSLLKKSSKNPE 438
Query: 367 EDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGY 426
E ++E TL++VM++F+Y+++KDVF+K+Y + LAKRL+ + SDDAE S+I KLK CG+
Sbjct: 439 EAELEDTLNQVMVVFKYIEDKDVFQKFYAKMLAKRLVHQNSASDDAEASMISKLKQACGF 498
Query: 427 QFTSKLEGMFTDMKTSQDTMQGFYA----SHP-DLGDGPTLTVQVLTTGSWPTQSSITCN 481
++TSKL+ MF D+ S+D + F S P DL ++QVL++GSWP Q S T
Sbjct: 499 EYTSKLQRMFQDIGVSKDLNEQFKKHLTNSEPLDL----DFSIQVLSSGSWPFQQSCTFA 554
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLML 541
LP E+ ++F ++Y H+GR+L+W + +L K ++ L ST+QM +L+
Sbjct: 555 LPSELERSYQRFTAFYASRHSGRKLTWLYQLSKGELVTNCFK-NRYTLQASTFQMAILLQ 613
Query: 542 FNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGE---DDAFSVN 598
+N D + +++ +T+I L + LQ L L VL E + D E D +
Sbjct: 614 YNTEDAYTVQQLTDSTQIKMDILAQVLQIL-LKSKLLVLEDENANVDEVELKPDTLIKLY 672
Query: 599 DKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLI 658
+ +K +V I V + E + E++ T + +EEDRK I+AAIVRIMK R++L H L+
Sbjct: 673 LGYKNKKLRVNI-NVPMKTEQKQEQETTHKNIEEDRKLLIQAAIVRIMKMRKVLKHQQLL 731
Query: 659 AEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
EV QL RF +KK I+ LIE+++LER D ++ Y YLA
Sbjct: 732 GEVLTQLSSRFKPRVPVIKKCIDILIEKEYLERVDGEKDTYSYLA 776
>CUL2_HUMAN (Q13617) Cullin homolog 2 (CUL-2)
Length = 745
Score = 345 bits (885), Expect = 2e-94
Identities = 215/704 (30%), Positives = 384/704 (54%), Gaps = 48/704 (6%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTF 95
G+RLY+ + H++ + K + ++ L +R W +++K + + Y++ F
Sbjct: 54 GERLYTETKIFLENHVRHLHKRVLESEE-QVLVMYHRYWEEYSKGADYMDCLYRYLNTQF 112
Query: 96 IPSAKKTP------------------VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSE 137
I K T + EL L++WR+ ++ ++ L+ LL ++++
Sbjct: 113 IKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMV--EPLQAILIRMLLREIKND 170
Query: 138 RTGEVIDRGIMRNITKMLMDLGP-------AVYGQDFEAHFLQVSAEFYQVESQRFIECC 190
R GE ++ ++ + + + Y + FE+ FL + E+Y+ E+ ++
Sbjct: 171 RGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQES 230
Query: 191 DCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNML 250
+C Y++K RL +E R Y+ P + K+ + +M+ +H L+ +H E N++
Sbjct: 231 NCSQYMEKVLGRLKDEEIRCRKYLHPSSYTKVIHECQQRMVADH-LQFLHAECH---NII 286
Query: 251 CDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDE 310
+K D+ MY L R V+ GL + + + HI + G + ++ + P FV+ +L+
Sbjct: 287 RQEKKNDMANMYVLLRAVSTGLPHMIQELQNHIHDEGLRATSNLTQENMPTLFVESVLEV 346
Query: 311 KDKYDKIINQAFNNDKSFQNALNSSFEYFINLNP-----RSPEFISLFVDDKLRKGLKGV 365
K+ ++IN N D+ F +AL+ + +N ++PE ++ + D+ L+K KG+
Sbjct: 347 HGKFVQLINTVLNGDQHFMSALDKALTSVVNYREPKSVCKAPELLAKYCDNLLKKSAKGM 406
Query: 366 NEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECG 425
E++VE L + +F+Y+ +KDVF+K+Y + LAKRL+ G ++S D+E ++I KLK CG
Sbjct: 407 TENEVEDRLTSFITVFKYIDDKDVFQKFYARMLAKRLIHGLSMSMDSEEAMINKLKQACG 466
Query: 426 YQFTSKLEGMFTDMKTSQDTMQGF--YASHPD--LGDGPTLTVQVLTTGSWP-TQS-SIT 479
Y+FTSKL M+TDM S D F + + D + G + + VL G+WP TQ+ S T
Sbjct: 467 YEFTSKLHRMYTDMSVSADLNNKFNNFIKNQDTVIDLGISFQIYVLQAGAWPLTQAPSST 526
Query: 480 CNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVL 539
+P E+ + F +Y +GR+L+W + ++K + G+ + V+TYQM VL
Sbjct: 527 FAIPQELEKSVQMFELFYSQHFSGRKLTWLHYLCTGEVKMNY-LGKPYVAMVTTYQMAVL 585
Query: 540 MLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVND 599
+ FNN++ +SYKE++ +T++ +L + ++SL VK ++ + +D+ + +FS+N
Sbjct: 586 LAFNNSETVSYKELQDSTQMNEKELTKTIKSLLDVK---MINHDSEKEDIDAESSFSLNM 642
Query: 600 KFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIA 659
FSSK K KI T QK++ E ++TR V+EDRK ++AAIVRIMK+R++L HN LI
Sbjct: 643 NFSSKRTKFKI-TTSMQKDTPQEMEQTRSAVDEDRKMYLQAAIVRIMKARKVLRHNALIQ 701
Query: 660 EVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
EV Q + RF + + +KK IE LI++ ++ER Y Y+A
Sbjct: 702 EVISQSRARFNPSISMIKKCIEVLIDKQYIERSQASADEYSYVA 745
>CUL2_MOUSE (Q9D4H8) Cullin homolog 2 (CUL-2)
Length = 745
Score = 340 bits (872), Expect = 8e-93
Identities = 212/704 (30%), Positives = 382/704 (54%), Gaps = 48/704 (6%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTF 95
G+RLY+ + +H++ + K + ++ L +R W +++K + + Y++ +
Sbjct: 54 GERLYAETKIFLESHVRHLYKRVLESEE-QVLVMYHRYWEEYSKGADYMDCLYRYLNTQY 112
Query: 96 IPSAKKTP------------------VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSE 137
I K T + EL L++WR+ ++ ++ L+ LL ++++
Sbjct: 113 IKKNKLTEADIQYGYGGVDMNEPLMEIGELALDMWRKLMV--EPLQNILIRMLLREIKND 170
Query: 138 RTGEVIDRGIMRNITKMLMDLGP-------AVYGQDFEAHFLQVSAEFYQVESQRFIECC 190
R GE ++ ++ + + + Y F + FL + E+Y+ E+ ++
Sbjct: 171 RGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQGIFVSPFLTETGEYYKQEASNLLQES 230
Query: 191 DCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNML 250
+C Y++K RL +E R Y+ P + K+ + +M+ +H L+ +H E ++
Sbjct: 231 NCSQYMEKVLGRLKDEEIRCRKYLHPSSYTKVIHECQQRMVADH-LQFLHSECHSIIQQ- 288
Query: 251 CDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDE 310
++ D+ MY L R V+ GL + E + HI + G + ++ + P FV+ +L+
Sbjct: 289 --ERKNDMANMYVLLRAVSSGLPHMIEELQKHIHDEGLRATSNLTQEHMPTLFVESVLEV 346
Query: 311 KDKYDKIINQAFNNDKSFQNALNSSFEYFINLNP-----RSPEFISLFVDDKLRKGLKGV 365
K+ ++IN N D+ F +AL+ + +N ++PE ++ + D+ L+K KG+
Sbjct: 347 HGKFVQLINTVLNGDQHFMSALDKALTSVVNYREPKSVCKAPELLAKYCDNLLKKSAKGM 406
Query: 366 NEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECG 425
E++VE L + +F+Y+ +KDVF+K+Y + LAKRL+ G ++S D+E ++I KLK CG
Sbjct: 407 TENEVEDKLTSFITVFKYIDDKDVFQKFYARMLAKRLIHGLSMSMDSEEAMINKLKQACG 466
Query: 426 YQFTSKLEGMFTDMKTSQDTMQGF--YASHPD--LGDGPTLTVQVLTTGSWP-TQS-SIT 479
Y+FTSKL M+TDM S D F + + D + G + + VL G+WP TQ+ S T
Sbjct: 467 YEFTSKLHRMYTDMSVSADLNNKFNNFIRNQDTVIDLGISFQIYVLQAGAWPLTQAPSST 526
Query: 480 CNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVL 539
+P E+ + F +Y +GR+L+W + ++K + G+ + V+TYQM VL
Sbjct: 527 FAIPQELEKSVQMFELFYSQHFSGRKLTWLHYLCTGEVKMNY-LGKPYVAMVTTYQMAVL 585
Query: 540 MLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVND 599
+ FNN++ +SYKE++ +T++ +L + ++SL VK ++ + +D+ + +FS+N
Sbjct: 586 LAFNNSETVSYKELQDSTQMNEKELTKTIKSLLDVK---MINHDSEKEDIDAESSFSLNM 642
Query: 600 KFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIA 659
FSSK K KI T QK++ E ++TR V+EDRK ++AAIVRIMK+R++L HN LI
Sbjct: 643 SFSSKRTKFKI-TTSMQKDTPQELEQTRSAVDEDRKMYLQAAIVRIMKARKVLRHNALIQ 701
Query: 660 EVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
EV Q + RF + + +KK IE LI++ ++ER Y Y+A
Sbjct: 702 EVISQSRARFNPSISMIKKCIEVLIDKQYIERSQASADEYSYVA 745
>CUL1_DROME (Q24311) Cullin homolog 1 (Lin-19 homolog protein)
Length = 774
Score = 331 bits (849), Expect = 3e-90
Identities = 215/710 (30%), Positives = 361/710 (50%), Gaps = 52/710 (7%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGS-FLEELNRKWNDHNKALQMIRDILMYMDRT 94
G +LY L + ++L E+ +A G L ++W + + ++ I Y++R
Sbjct: 75 GKKLYDRLEQFLKSYLSELLTKFKAISGEEVLLSRYTKQWKSYQFSSTVLDGICNYLNRN 134
Query: 95 FIPSAKKT------PVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIM 148
++ + ++ L L W+ + + + +L+ ++ ER G++I+R ++
Sbjct: 135 WVKRECEEGQKGIYKIYRLALVAWKGHLF--QVLNEPVTKAVLKSIEEERQGKLINRSLV 192
Query: 149 RNITKMLMDLG------------PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYL 196
R++ + ++L +VY Q+FE F+ ++ FY+ ES F+ +YL
Sbjct: 193 RDVIECYVELSFNEEDTDAEQQKLSVYKQNFENKFIADTSAFYEKESDAFLSTNTVTEYL 252
Query: 197 KKAERRLNEEMDRVG--------HYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVN 248
K E RL EE RV Y+ T + E +IE H L++ H E L+N
Sbjct: 253 KHVENRLEEETQRVRGFNSKNGLSYLHETTADVLKSTCEEVLIEKH-LKIFHTEFQNLLN 311
Query: 249 MLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTD---PERLKDPVEFVQ 305
D+ +DL RMY+L + L ++ ++ HI G + + + DP +VQ
Sbjct: 312 A---DRNDDLKRMYSLVALSSKNLTDLKSILENHILHQGTEAIAKCCTTDAANDPKTYVQ 368
Query: 306 RLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLN--------PRSPEFISLFVDDK 357
+LD KY+ ++ AFNND F AL+ + FIN N +SPE ++ + D
Sbjct: 369 TILDVHKKYNALVLTAFNNDNGFVAALDKACGKFINSNVVTIANSASKSPELLAKYCDLL 428
Query: 358 LRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLI 417
L+K K + ++E L++VM++F+Y+++KDVF+KYY + LAKRL++ + SDDAE +I
Sbjct: 429 LKKSSKNPEDKELEDNLNQVMVVFKYIEDKDVFQKYYSKMLAKRLVNHTSASDDAEAMMI 488
Query: 418 VKLKTECGYQFTSKLEGMFTDMKTSQDTMQGF--YASHPDLGDGPTLTVQVLTTGSWPTQ 475
KLK CGY++T KL+ MF D+ S+D F Y + +L ++VL++GSWP Q
Sbjct: 489 SKLKQTCGYEYTVKLQRMFQDIGVSKDLNSYFKQYLAEKNLTMEIDFGIEVLSSGSWPFQ 548
Query: 476 SSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNM--GFADLKATFGKGQKHELNVST 533
S LP E+ +F +Y H+GR+L+W M G + + L ST
Sbjct: 549 LSNNFLLPSELERSVRQFNEFYAARHSGRKLNWLYQMCKGELIMNVNRNNSSTYTLQAST 608
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDD 593
+QM VL+ FN+ + ++++ T+ +L + LQ L K VL + +
Sbjct: 609 FQMSVLLQFNDQLSFTVQQLQDNTQTQQENLIQVLQILLKAK---VLTSSDNENSLTPES 665
Query: 594 AFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLD 653
+ + +K ++ I + + E + E++ + +EEDRK I+AAIVRIMK R+ L+
Sbjct: 666 TVELFLDYKNKKRRININQPL-KTELKVEQETVHKHIEEDRKLLIQAAIVRIMKMRKRLN 724
Query: 654 HNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
H NLI+EV QL RF +KK I+ LIE+++LER + + Y YLA
Sbjct: 725 HTNLISEVLNQLSTRFKPKVPVIKKCIDILIEKEYLERMEGHKDTYSYLA 774
>CUL1_SCHPO (O13790) Cullin 1 homolog (Cul-1) (Cell division control
53 homolog)
Length = 767
Score = 320 bits (819), Expect = 1e-86
Identities = 215/707 (30%), Positives = 353/707 (49%), Gaps = 43/707 (6%)
Query: 26 NAYNMVLHKFGDRLYSGLVATMTAHLKEIAKS-IEAAQGGSFLEELNRKWNDHNKALQMI 84
N + + G+ LY+ LV + +L + K I L + W + + I
Sbjct: 69 NFNDQTANVLGEALYNNLVLYLEEYLARLRKECISQTNHEEQLAAYAKYWTRFTTSARFI 128
Query: 85 RDILMYMDRTFIPSAKKTP------VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSER 138
+ Y++R ++ + ++ L L W V + IR LL LL + +R
Sbjct: 129 NHLFGYLNRYWVKLKNRFTETLVYDIYTLCLVSWHHHVF--SHIRDSLLQNLLYMFTKKR 186
Query: 139 TGEVIDRGIMR----NITKMLMDLGPAV------YGQDFEAHFLQVSAEFYQVESQRFIE 188
E D + +IT + D Y FE +F++ + FY ES ++
Sbjct: 187 LYEPTDMKYVEVCVDSITSLSFDKTDMTKPNLSSYKTFFETNFIENTKNFYAKESSEYLA 246
Query: 189 CCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVN 248
DYLKKAE RL EE + V Y+ T K + + E +I H ++H + + +++
Sbjct: 247 SHSITDYLKKAEIRLAEEEELVRLYLHESTLKPLLEATEDVLIAQHE-EVLHNDFARMLD 305
Query: 249 MLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTD------PERLKDPVE 302
C ED+ RMY L R +GL +R+ ++ SG V E DP E
Sbjct: 306 QNCS---EDIIRMYRLMSRTPNGLQPLRQTFEEFVKRSGFAAVAKIVPQVGGEADVDPKE 362
Query: 303 FVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLN-------PRSPEFISLFVD 355
+++ LL +++N AF+ D F +L+++F +N N RSPE ++ + D
Sbjct: 363 YMEMLLSTYKASKELVNTAFHGDTDFTKSLDTAFRELVNRNVVCQRSSSRSPELLAKYAD 422
Query: 356 DKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERS 415
LRK K V+ DDVE L ++++FRY+++KDVF+ +Y + LAKRL++G + S DAE S
Sbjct: 423 SILRKSNKNVDIDDVEDCLSSIIIIFRYVEDKDVFQNFYTKLLAKRLVNGTSNSQDAESS 482
Query: 416 LIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWP-T 474
++ KLK CG+++TSKL+ MF D+ SQ+ + F+ + VL T WP +
Sbjct: 483 MLSKLKEVCGFEYTSKLQRMFQDISLSQEITEAFWQLPQSRAGNIDFSALVLGTSFWPLS 542
Query: 475 QSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFG--KGQKHELNVS 532
+++ +LP E+ L E F++YY H GR+LSW ++ ++KA + VS
Sbjct: 543 PNNVNFHLPEELVPLYEGFQNYYYSCHNGRKLSWLFHLSKGEIKARINPQTNVTYVFQVS 602
Query: 533 TYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGED 592
TYQM VL+L+N+ D +Y+E+ + T + L L K VL K +
Sbjct: 603 TYQMGVLLLYNHRDSYTYEELAKITGLSTDFLTGILNIFLKAK---VLLLGDNDKLGDPN 659
Query: 593 DAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLL 652
+ +N+ F K +V++ + + E + E ET + +EEDRK +++AIVRIMK+RR L
Sbjct: 660 STYKINENFRMKKIRVQLNLPI-RSEQKQESLETHKTIEEDRKLLLQSAIVRIMKARRTL 718
Query: 653 DHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMY 699
H L+ E Q++ RF +++K+ I+ LIE+++LER D +Y
Sbjct: 719 KHVVLVKETIDQIKSRFTPKVSDIKQCIDMLIEKEYLERQGRDEYIY 765
>CUL1_CAEEL (Q17389) Cullin 1 (Abnormal cell lineage 19 protein)
Length = 780
Score = 316 bits (809), Expect = 2e-85
Identities = 210/713 (29%), Positives = 364/713 (50%), Gaps = 58/713 (8%)
Query: 36 GDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTF 95
G +Y + + A++ + + G L+ +W + + +++ I Y++R +
Sbjct: 81 GHEMYQRVEEYVKAYVIAVCEKGAELSGEDLLKYYTTEWENFRISSKVMDGIFAYLNRHW 140
Query: 96 IPSAKKTP------VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMR 149
I V+ L L +W+ ++ N ++ ++++ +LEL++SERTG +I+ +
Sbjct: 141 IRRELDEGHENIYMVYTLALVVWKRNLF--NDLKDKVIDAMLELIRSERTGSMINSRYIS 198
Query: 150 NITKMLMDLGP----------------AVYGQDFEAHFLQVSAEFYQVESQRFIECC-DC 192
+ + L++LG AVY + FE FL+ + FY E+ F+ +
Sbjct: 199 GVVECLVELGVDDSETDAKKDAETKKLAVYKEFFEVKFLEATRGFYTQEAANFLSNGGNV 258
Query: 193 GDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCD 252
DY+ K E RLN+E DR Y++ T+ + E+ +I N L + GL L D
Sbjct: 259 TDYMIKVETRLNQEDDRCQLYLNSSTKTPLATCCESVLISNQ-LDFLQRHFGGL---LVD 314
Query: 253 DKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLV--TDPERLKDPVEFVQRLLDE 310
+ +DL RM+ L RV +GL ++R+ + HI + G Q + E D +V+ LL+
Sbjct: 315 KRDDDLSRMFKLCDRVPNGLDELRKSLENHIAKEGHQALERVAMEAATDAKLYVKTLLEV 374
Query: 311 KDKYDKIINQAFNNDKSFQNALNSSFEYFINLNP------------RSPEFISLFVDDKL 358
++Y ++N++F N+ F +L+ + FIN N +S E ++ + D L
Sbjct: 375 HERYQSLVNRSFKNEPGFMQSLDKAATSFINNNAVTKRAPPQAQLTKSAELLARYCDQLL 434
Query: 359 RKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIV 418
RK K +E ++E K+M++F+Y+ +KDVF K+Y + +KRL+S + SD+AE + I
Sbjct: 435 RKSSKMPDEAELEELQTKIMVVFKYIDDKDVFSKFYTKMFSKRLISELSASDEAEANFIT 494
Query: 419 KLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHPDL--GDGPTLTVQVLTTGSWPTQS 476
KLK+ CGY++T++L M D + S+D F D+ V VL++GSWPT
Sbjct: 495 KLKSMCGYEYTARLSKMVNDTQVSKDLTADFKEKKADMLGQKSVEFNVLVLSSGSWPTFP 554
Query: 477 SITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQM 536
+ LP ++S E F +Y GRRL+W + ++ +T +K+ +T QM
Sbjct: 555 TTPITLPQQLSKTIEIFGQFYNEKFNGRRLTWVYSQSRGEITST-AFPKKYVFTATTAQM 613
Query: 537 CVLMLFNNADKLSYKEIEQAT---EIPAPDLKRCLQSLALVKGRNVLRKE---PMSKDVG 590
C ++LFN D + ++I AT E AP + L ++K L+KE PM+ V
Sbjct: 614 CTMLLFNEQDSYTVEQIAAATKMDEKSAPAIVGSLIKNLVLKADTELQKEDEVPMTATV- 672
Query: 591 EDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRR 650
S+N + +K +V + ++++ + + ++ VEEDRK I A IVRIMK+R+
Sbjct: 673 -----SLNKAYMNKKVRVDLSKFTMKQDAVRDTENVQKNVEEDRKSVISACIVRIMKTRK 727
Query: 651 LLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ H L+ EV QL RF +K+ I SLIE++++ R + + +Y YLA
Sbjct: 728 RVQHQQLMTEVITQLSGRFKPKVEMIKRCIGSLIEKEYMLRTEGQKDLYEYLA 780
>CUL4_CAEEL (Q17392) Cullin 4
Length = 840
Score = 287 bits (735), Expect = 6e-77
Identities = 208/699 (29%), Positives = 364/699 (51%), Gaps = 53/699 (7%)
Query: 39 LYSGLVATMTAHLKEIAKSIEAAQG--------GSFLEELNRKWNDHNKALQMIRDILMY 90
LY LVA + K + +S+ A + +LE+ + W + + +IR+I ++
Sbjct: 161 LYDRLVAIVVQFAKSLKESLNAVEQVPLAEDNCEQYLEKFGQIWQAYPVKINLIRNIFLH 220
Query: 91 MDRTFIPSAKKT--PVHELGLNLWRESV---IYSNQIRTRLLNTLLELVQSERTGEVIDR 145
+DR + + P+ E + +++++ I+ T+L N L +Q +++ R
Sbjct: 221 LDRIALGATDTEILPLWECFMQIFQKTFFPNIFKEFKATKLFNALYMAMQ-----KIMQR 275
Query: 146 GIMRNITKMLMDLGPAVY-GQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLN 204
+ + + L+D+ V+ ++F + E Y E + +C DY++ E ++N
Sbjct: 276 YPVDSPLRSLVDMLQTVHVSEEFAKFLISQLREHYNNERIDKVPKMNCNDYMEYCEDQIN 335
Query: 205 EEMDRVG-HYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
V ++ +P K + V +I+ + ++ + L++ D D+GRM+N
Sbjct: 336 RYSQLVKVNFDEPSALKDVQATVTNCLIQQAIPEILTHDFDDLID---SDNISDIGRMFN 392
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLV-TDPERLKDPVEFVQRLLDEKDKYDKIINQAF 322
L R+ G ++R + ++++ G++L+ T P+ + V LL K K D I+ +F
Sbjct: 393 LCRQCVGGEDEVRTQFSKYLKKRGEKLIATCPDE-----DLVSELLAFKKKVDFIMTGSF 447
Query: 323 ---NNDKSFQNALNSSFEYFINLN-PRSPEFISLFVDDKLRKGLKGVNEDD-VEVTLDKV 377
N+ + L+ +FE F+N RS E IS L K V++D ++ +D+
Sbjct: 448 KSANDPVKMRQCLSDAFESFVNKQVDRSAELISKHFHTLLHSSNKNVSDDTTLDQMVDEA 507
Query: 378 MMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFT 437
++LFRYL+ KDVFE YYK+ LAKRL ++ S DAE+ ++ KLKTECG FT KLEGMF
Sbjct: 508 IVLFRYLRGKDVFEAYYKRGLAKRLFLERSASVDAEKMVLCKLKTECGSAFTYKLEGMFK 567
Query: 438 DMKTSQDTMQGF--YASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRS 495
DM S++ + F Y H + + T +V+T WPT + N+P E+ ++
Sbjct: 568 DMDASENYGRLFNQYLEHMN-KEKANFTARVITPEYWPTYDTYEINIPKEMRDTLTDYQD 626
Query: 496 YYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQ 555
+Y H R + W + A + A+F G K EL + YQ +L+LFN + + EI +
Sbjct: 627 FYRVQHGNRNVKWHHGLASAVISASFRPGCKKELIATMYQTVILLLFNKCETWTVAEIVE 686
Query: 556 ATEIPAPDLKRCLQSLALVKGRN---VLR--------KEPMSKDVGEDDAFSVNDKFSSK 604
T+I +++ LAL+ GR+ VL+ K+ + + +++ F VN KF+ K
Sbjct: 687 HTKI--LEIEVVKNVLALLGGRDKPKVLQRVEGGGSEKKEGTVENLKNEKFVVNSKFTEK 744
Query: 605 LYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQ 664
+V+I V K + E +E ++ V DR+ +I+AA+VRIMK+R+ L+H L+ E+ +
Sbjct: 745 RCRVRIAQVNI-KTAVEETKEVKEEVNSDRQYKIDAAVVRIMKARKQLNHQTLMTELLQ- 802
Query: 665 LQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
QLRF + ++KKR+ESLIER+++ RD + Y Y+A
Sbjct: 803 -QLRFPVSTADIKKRLESLIEREYISRDPEEASSYNYVA 840
>CUL6_CAEEL (Q21346) Cullin 6
Length = 729
Score = 283 bits (724), Expect = 1e-75
Identities = 199/690 (28%), Positives = 345/690 (49%), Gaps = 44/690 (6%)
Query: 39 LYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTFIP- 97
LY L + ++ I I A G L + +W++ + ++ I Y++R F+
Sbjct: 59 LYKQLENYIRTYVIAIRDRISACSGDELLGKCTIEWDNFKFSTRICNCIFQYLNRNFVSK 118
Query: 98 -----SAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNIT 152
+ + +++L L++W+ + +++T ++ +LEL+ ER G I+ + ++
Sbjct: 119 KVEDKNGEIVEIYKLALDIWKAEFFDNFKVKT--IDAILELILLERCGSTINSTHISSVV 176
Query: 153 KMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGH 212
+ L +L +Y FE FL + FY+ E E +Y+ E RL +E R
Sbjct: 177 ECLTELD--IYKVSFEPQFLDATKLFYKQEVLNSKETVI--EYMITVENRLFQEEYRSRR 232
Query: 213 YMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGL 272
Y+ P T + E+ +I + L+ +H E L+ D E L RMY+L RRV GL
Sbjct: 233 YLGPSTNDLLIDSCESILISDR-LKFLHSEFERLLEARKD---EHLTRMYSLCRRVTHGL 288
Query: 273 LKIREVMTLHIRESG----KQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNNDKSF 328
+R + I + G ++L D P E++ +LL+ + Y +IN+AF+ + F
Sbjct: 289 EDLRVYLEKRILKEGHETLQRLAKDSGLKTTPKEYITKLLEVHEIYFNLINKAFDRNALF 348
Query: 329 QNALNSSFEYFINLNP------------RSPEFISLFVDDKLRKGLKGVNEDDVEVTLDK 376
+L+ + + FI N RS ++++ + D L+K K +E LDK
Sbjct: 349 MQSLDKASKDFIEANAVTMLAPEKHRSTRSADYLARYCDQLLKKNSKVQDE----TALDK 404
Query: 377 VMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMF 436
+ + +Y+ EKDVF+ YY+ ++R+++ + SDDAE I L G ++T L M
Sbjct: 405 ALTVLKYISEKDVFQLYYQNWFSERIINNSSASDDAEEKFITNLTATEGLEYTRNLVKMV 464
Query: 437 TDMKTSQDTMQGFYASHPDLGDGPTLTVQVL--TTGSWPTQSSITCNLPVEISALCEKFR 494
D K S+D F D+ ++ V+ TTG+WP+ I LP E+S + ++F
Sbjct: 465 EDAKISKDLTTEF----KDIKTEKSIDFNVILQTTGAWPSLDQIKIILPRELSTILKEFD 520
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIE 554
++Y +H GRRL+W + ++ + + +K+ V+ Q+C L LFN D + ++I
Sbjct: 521 TFYNASHNGRRLNWAYSQCRGEVNSKAFE-KKYVFIVTASQLCTLYLFNEQDSFTIEQIS 579
Query: 555 QATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAF-SVNDKFSSKLYKVKIGTV 613
+A E+ A + SL V ++ + KD DA S+N K+++K +V + T
Sbjct: 580 KAIEMTAKSTSAIVGSLNPVIDPVLVVDKGNEKDGYPPDAVVSLNTKYANKKVRVDLTTA 639
Query: 614 VAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANP 673
+ + ++ E + VE DRK +I+A IVRIMK+R+ L H LI E+ QL+ RF N
Sbjct: 640 IKKATADRETDAVQNTVESDRKYEIKACIVRIMKTRKSLTHTLLINEIISQLKSRFTPNV 699
Query: 674 TEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+K IE LIE+ ++ R +N+ +Y YLA
Sbjct: 700 QMIKICIEILIEQLYIRRSENEHNVYEYLA 729
>CUL4_SCHPO (O14122) Cullin 4 homolog (Cul-4)
Length = 734
Score = 283 bits (723), Expect = 1e-75
Identities = 196/640 (30%), Positives = 331/640 (51%), Gaps = 28/640 (4%)
Query: 74 WNDHNKALQMIRDILMYMDRTFIPSAKKTP-VHELGLNLWRESVIYSNQIRTRLLNTLLE 132
WN + ++++++I YMD+TF+ P + EL L+L+RE ++ I+ LN+LL+
Sbjct: 111 WNKWLERVEIVQNIFYYMDKTFLSHHPDYPTIEELSLSLFREKLMAVKNIQIPFLNSLLQ 170
Query: 133 LVQSERTGEVIDRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDC 192
++ + + D +++ MLM +Y F +L + + FY ES + I+
Sbjct: 171 SFENLHSSKSTDHAYLQDA--MLMLHRTEMYSSVFVPMYLVMLSRFYDTESSQKIQELPL 228
Query: 193 GDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCD 252
+YL+ A L E V + +K I + V+ +I +H+ L G+ +
Sbjct: 229 EEYLEYAMSSLEREDAYVEKFDIVRDKKSIRETVQRCLITSHLDTL----TKGISQFIEK 284
Query: 253 DKYEDLGRMYNL--FRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDE 310
+Y L F + L++ + + G +LV D KD VQ LL
Sbjct: 285 RDAHSCKLLYALLQFNHETEYLIQPWSDCLVDV---GFKLVNDES--KDDT-LVQELLSF 338
Query: 311 KDKYDKIINQAFNNDKSFQNALNSSFEYFINLNPRSPE-----FISLFVDDKLRKGLKGV 365
+++++F +D++ A+ +FE FIN S I+ ++D LR G +
Sbjct: 339 HKFLQVVVDESFLHDETLSYAMRKAFETFINGAKGSQREAPARLIAKYIDYLLRVGEQAS 398
Query: 366 NEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECG 425
++ +++ LFRY+ KD+FE YYK +AKRLL K+ S E L+ LK CG
Sbjct: 399 GGKPLKEVFSEILDLFRYIASKDIFEAYYKLDIAKRLLLNKSASAQNELMLLDMLKKTCG 458
Query: 426 YQFTSKLEGMFTDMKTSQDTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVE 485
QFT LEGMF D+ S++ F S L V VL+ WP+ LP +
Sbjct: 459 SQFTHSLEGMFRDVNISKEFTSSFRHSKAAHNLHRDLYVNVLSQAYWPSYPESHIRLPDD 518
Query: 486 ISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNN- 544
+ + F +YL G+++SW ++G +KA F G K EL++S +Q CVL+ FNN
Sbjct: 519 MQQDLDCFEKFYLSKQVGKKISWYASLGHCIVKARFPLGNK-ELSISLFQACVLLQFNNC 577
Query: 545 --ADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFS 602
+ +SY++++++TE+ DL R LQSL+ + R ++ P SK D F VN+KF+
Sbjct: 578 LGGEGISYQDLKKSTELSDIDLTRTLQSLSCARIRPLV-MVPKSKKPSPDTMFYVNEKFT 636
Query: 603 SKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVT 662
KLY+VKI + KE E + +++V DR+ +++A+IVR+MK + + H++L+ V
Sbjct: 637 DKLYRVKINQIYL-KEERQENSDVQEQVVRDRQFELQASIVRVMKQKEKMKHDDLVQYVI 695
Query: 663 KQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYL 702
++ R + ++VK IE L+E+++LER+DND +Y Y+
Sbjct: 696 NNVKDRGIPLVSDVKTAIEKLLEKEYLEREDND--IYTYV 733
>CUL2_CAEEL (Q17390) Cullin 2
Length = 776
Score = 275 bits (702), Expect = 4e-73
Identities = 193/736 (26%), Positives = 360/736 (48%), Gaps = 84/736 (11%)
Query: 37 DRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDILMYMDRTFI 96
+RLY+ + A + H+++ + I L+E ++ W ++ I + Y+++ F+
Sbjct: 56 ERLYNEVKACIQEHVRQKRQDIVDVDPDLLLQEYHKMWRVFHEGAIFIHRLFGYLNKQFV 115
Query: 97 PSAKKTP---------------VHELG---LNLWRESVIYSNQIRTRLLNTLLELVQSER 138
+ T V E+G L +W+E ++ + I +L+ LL + ++R
Sbjct: 116 KQKRCTDLDNFAQYAAFLQIPDVKEIGCLALEIWKEDLVKT--ILPQLVKLLLIAIDNDR 173
Query: 139 TGEVID-----RGIMRNITKML---MDLGPA-------------VYGQDFEAHFLQVSAE 177
G G++ + KM D+ PA Y + FE L + +
Sbjct: 174 KGNFPHIANEVSGVINSFVKMEETDFDVVPAEGARYKARESTTAFYQESFEKPLLTDTEQ 233
Query: 178 FYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLR 237
+Y +Q+ + C +Y+++ L +E R Y+ + +K+ + + MI+ H +
Sbjct: 234 YYSALAQKMLTDLSCSEYMEQVIVLLEQEEMRAKKYLHESSVEKVITLCQKVMIKAHKDK 293
Query: 238 LIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERL 297
L ++ +++ +++ +DL MY L + + GL + + ++++ G + V+
Sbjct: 294 L----HAVCHDLITNEENKDLRNMYRLLKPIQAGLSVMVKEFEEYVKKKGLEAVSRLTGE 349
Query: 298 KDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLN------PRSPEFIS 351
P +FV+ +L +K++ + F +D F + L+ + + +N P++ E ++
Sbjct: 350 NVPQQFVENVLRVYNKFNDMKTAVFMDDGEFSSGLDKALQGVVNSKEPGQSVPKASERLA 409
Query: 352 LFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDD 411
+ D L+K KG++E D+E LD +++FRY+++KD+F+K+Y + LA RL++ ++S D
Sbjct: 410 RYTDGLLKKSTKGLSETDLEAKLDSAIVIFRYIEDKDIFQKFYSKMLANRLIASTSISMD 469
Query: 412 AERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGF-------YASHPDLGDGPTLTV 464
AE +I KLK CGY+FTSKL MFTD+ SQ+ F PD+ PT T+
Sbjct: 470 AEELMINKLKQACGYEFTSKLSRMFTDIGLSQELSNNFDKHIADIKTVQPDVKFVPTQTM 529
Query: 465 QVLTTGSWP--------------TQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQT 510
+L GSWP Q +LP + + ++F +Y G H GR+L+W
Sbjct: 530 -ILQAGSWPLNAPQLSTNSNNQTAQDVANFHLPRILQPVIQEFEKFYTGKHNGRKLTWLF 588
Query: 511 NMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQS 570
NM D++ T+ +++ + YQM L+ F D + K+I + + L + +++
Sbjct: 589 NMSQGDVRLTY-LDKQYVAQMYVYQMAALLCFERRDAILVKDIGEEIGVSGDYLLKTIRT 647
Query: 571 LALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQ-- 628
+ +V +++ D +N +SK K ++ K E E++
Sbjct: 648 IL-----DVTLLTCDDQNLTADSLVRLNMSMTSKRMKFRLQAPQVNKAVEKEQEAVANTF 702
Query: 629 RVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDF 688
+V +DRK +E AIVRIMK+R++L HN L+ E+ Q + RF + +KK IE LIE+ +
Sbjct: 703 QVSQDRKYYMECAIVRIMKTRKVLKHNALVTEIMDQTKGRFSPDVPFIKKSIEDLIEKMY 762
Query: 689 LER-DDNDRKMYRYLA 703
++R D ND Y+YLA
Sbjct: 763 IQRTDQNDE--YQYLA 776
>CUL5_HUMAN (Q93034) Cullin homolog 5 (CUL-5) (Vasopressin-activated
calcium-mobilizing receptor) (VACM-1)
Length = 780
Score = 229 bits (583), Expect = 2e-59
Identities = 182/657 (27%), Positives = 332/657 (49%), Gaps = 68/657 (10%)
Query: 104 VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGP--- 160
V +L L+ W ES I+SN I+ RL ++ ++LV +ER GE D ++ + + ++L
Sbjct: 135 VRKLMLDTWNES-IFSN-IKNRLQDSAMKLVHAERLGEAFDSQLVIGVRESYVNLCSNPE 192
Query: 161 ---AVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPE 217
+Y +FE +L + FY+ ++ +++ +Y+K A+ +L EE R Y+ E
Sbjct: 193 DKLQIYRDNFEKAYLDSTERFYRTQAPSYLQQNGVQNYMKYADAKLKEEEKRALRYL--E 250
Query: 218 TEKKINKVVE-----TQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGL 272
T ++ N V + I E G++ ++ E L M++L +V +G+
Sbjct: 251 TRRECNSVEALMECCVNALVTSFKETILAECQGMIKR---NETEKLHLMFSLMDKVPNGI 307
Query: 273 LKIREVMTLHIRESG-KQLVTDPERLK-DPVEFVQRLLDEKDKYDKIINQAFNNDKSFQN 330
+ + + HI +G +V E + D ++V++LL +++ K++ +AF +D F
Sbjct: 308 EPMLKDLEEHIISAGLADMVAAAETITTDSEKYVEQLLTLFNRFSKLVKEAFQDDPRFLT 367
Query: 331 ALNSSFEYFIN-------------------LNPRS--PEFISLFVDDKLRKG--LKGVNE 367
A + +++ +N P S PE ++ + D LRK K +
Sbjct: 368 ARDKAYKAVVNDATIFKLELPLKQKGVGLKTQPESKCPELLANYCDMLLRKTPLSKKLTS 427
Query: 368 DDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGY- 426
+++E L +V+++ +Y+Q KDVF +Y+K HL +RL+ + + E +++ L+ E G
Sbjct: 428 EEIEAKLKEVLLVLKYVQNKDVFMRYHKAHLTRRLILDISADSEIEENMVEWLR-EVGMP 486
Query: 427 -QFTSKLEGMFTDMKTSQDTMQGFYASHPDLG---DGPTLTVQVLTTGSWPTQSS-ITCN 481
+ +KL MF D+K S+D Q F H + ++ +++L G+W S + +
Sbjct: 487 ADYVNKLARMFQDIKVSEDLNQAFKEMHKNNKLALPADSVNIKILNAGAWSRSSEKVFVS 546
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNM--GFADLKATFGKGQKHELNVSTYQMCVL 539
LP E+ L + +Y H+GR+L W M G K G+ ++L V+T+Q+ VL
Sbjct: 547 LPTELEDLIPEVEEFYKKNHSGRKLHWHHLMSNGIITFKNEVGQ---YDLEVTTFQLAVL 603
Query: 540 MLFNNA--DKLSYKEIEQATEIPAPDLKRCLQSLALVKG--RNVLRKEPM---SKDVGED 592
+N +K+S++ ++ ATE+P +L+R L SL R VL EP KD E
Sbjct: 604 FAWNQRPREKISFENLKLATELPDAELRRTLWSLVAFPKLKRQVLLYEPQVNSPKDFTEG 663
Query: 593 DAFSVNDKFS----SKLYKV-KIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIM 646
FSVN +FS +K+ K KI + Q +E ++E + + + R + + AI++IM
Sbjct: 664 TLFSVNQEFSLIKNAKVQKRGKINLIGRLQLTTERMREEENEGIVQLRILRTQEAIIQIM 723
Query: 647 KSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
K R+ + + L E+ + L+ FL +K++IE LIE ++ RD++D + Y+A
Sbjct: 724 KMRKKISNAQLQTELVEILKNMFLPQKKMIKEQIEWLIEHKYIRRDESDINTFIYMA 780
>CUL5_RAT (Q9JJ31) Cullin homolog 5 (CUL-5) (Vasopressin-activated
calcium-mobilizing receptor) (VACM-1)
Length = 780
Score = 225 bits (573), Expect = 4e-58
Identities = 181/657 (27%), Positives = 329/657 (49%), Gaps = 68/657 (10%)
Query: 104 VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGP--- 160
V +L L+ W ES I+SN I+ RL ++ ++LV +ER GE D ++ + + ++L
Sbjct: 135 VRKLMLDTWNES-IFSN-IKNRLQDSAMKLVHAERLGEAFDSQLVIGVRESYVNLCSNPE 192
Query: 161 ---AVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPE 217
+Y +FE +L + FY+ ++ +++ +Y+K A+ +L EE R Y+ E
Sbjct: 193 DKLQIYRDNFEKAYLDSTERFYRTQAPSYLQQNGVQNYMKYADAKLKEEEKRALRYL--E 250
Query: 218 TEKKINKVVE-----TQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGL 272
T ++ N V + I E G++ ++ E L M++L +V G+
Sbjct: 251 TRRECNSVEALMECCVNALVTSFKETILAECQGMIKR---NETEKLHLMFSLMDKVPGGI 307
Query: 273 LKIREVMTLHIRESG-KQLVTDPERLK-DPVEFVQRLLDEKDKYDKIINQAFNNDKSFQN 330
+ + + HI +G +V E + D ++V++LL +++ K++ +AF +D F
Sbjct: 308 EPMLKDLEEHIISAGLADMVAAAETITTDSEKYVEQLLTLFNRFSKLVKEAFQDDPRFLT 367
Query: 331 ALNSSFEYFIN-------------------LNPRS--PEFISLFVDDKLRKG--LKGVNE 367
A + +++ +N P S PE ++ + D LRK K +
Sbjct: 368 ARDKAYKAVVNDATIFKLELPLKQKGVGLKTQPESKCPELLANYCDMLLRKTPLSKKLTS 427
Query: 368 DDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGY- 426
+++E L +V+++ +Y+Q KDVF +Y+K HL +RL+ + + E +++ L+ E G
Sbjct: 428 EEIEAKLKEVLLVLKYVQNKDVFMRYHKAHLTRRLILDISADSEIEENMVEWLR-EVGMP 486
Query: 427 -QFTSKLEGMFTDMKTSQDTMQGFYASHPDLG---DGPTLTVQVLTTGSWPTQSS-ITCN 481
+ +KL MF D+K S+D Q F H + ++ +++L G+W S + +
Sbjct: 487 ADYVNKLARMFQDIKVSEDLNQAFKEMHKNNKLALPADSVNIKILNAGAWSRSSEKVFVS 546
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNM--GFADLKATFGKGQKHELNVSTYQMCVL 539
LP E+ L + +Y H+GR+L W M G K G+ ++L V+T+Q+ VL
Sbjct: 547 LPTELEDLIPEVEEFYKKNHSGRKLHWHHLMSNGIITFKNEVGQ---YDLEVTTFQLAVL 603
Query: 540 MLFNNA--DKLSYKEIEQATEIPAPDLKRCLQSLALVKG--RNVLRKEPM---SKDVGED 592
+N +K+S++ ++ ATE+P +L+R L SL R VL +P KD E
Sbjct: 604 FAWNQRPREKISFENLKLATELPDAELRRTLWSLVAFPKLKRQVLLYDPQVNSPKDFTEG 663
Query: 593 DAFSVNDKFS----SKLYKV-KIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIM 646
FSVN FS +K+ K KI + Q +E ++E + + + R + + AI++IM
Sbjct: 664 TLFSVNQDFSLIKNAKVQKRGKINLIGRLQLTTERMREEENEGIVQLRILRTQEAIIQIM 723
Query: 647 KSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
K R+ + + L E+ + L+ FL +K++IE LIE ++ RD+ D + Y+A
Sbjct: 724 KMRKKISNAQLQTELVEILKNMFLPQKKMIKEQIEWLIEHKYIRRDEADINTFIYMA 780
>CUL5_RABIT (Q29425) Cullin homolog 5 (CUL-5) (Vasopressin-activated
calcium-mobilizing receptor) (VACM-1)
Length = 780
Score = 224 bits (571), Expect = 6e-58
Identities = 180/657 (27%), Positives = 329/657 (49%), Gaps = 68/657 (10%)
Query: 104 VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGP--- 160
V +L L+ W ES I+SN I+ RL ++ ++LV +ER GE D ++ + + ++L
Sbjct: 135 VRKLMLDTWNES-IFSN-IKNRLQDSAMKLVHAERLGEAFDSQLVIGVRESYVNLCSNPE 192
Query: 161 ---AVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPE 217
+Y +FE +L + FY+ ++ +++ +Y+K A+ +L EE R Y+ E
Sbjct: 193 DKLQIYRDNFEKAYLDSTERFYRTQAPSYLQQNGVQNYMKYADAKLKEEEKRALRYL--E 250
Query: 218 TEKKINKVVE-----TQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGL 272
T ++ N V + I E G++ ++ E L M++L +V +G+
Sbjct: 251 TRRECNSVEALMECCVNALVTSFKETILAECQGMIKR---NETEKLHLMFSLMDKVPNGI 307
Query: 273 LKIREVMTLHIRESG-KQLVTDPERLK-DPVEFVQRLLDEKDKYDKIINQAFNNDKSFQN 330
+ + + HI +G +V E + D ++V++LL +++ K++ +AF +D F
Sbjct: 308 EPMLKDLEEHIISAGLADMVAAAETITTDSEKYVEQLLTLFNRFSKLVKEAFQDDPRFLT 367
Query: 331 ALNSSFEYFIN-------------------LNPRS--PEFISLFVDDKLRKG--LKGVNE 367
A + +++ +N P S PE ++ + D LRK K +
Sbjct: 368 ARDKAYKAVVNDATIFKLELPLKQKGVGLKTQPESKCPELLANYCDMLLRKTPLSKKLTS 427
Query: 368 DDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGY- 426
+++E L +V+++ +Y+Q KDVF +Y+K HL +RL+ + + E +++ L+ E G
Sbjct: 428 EEIEAKLKEVLLVLKYVQNKDVFMRYHKAHLTRRLILDISADSEIEENMVEWLR-EVGMP 486
Query: 427 -QFTSKLEGMFTDMKTSQDTMQGFYASHPD---LGDGPTLTVQVLTTGSWPTQS-SITCN 481
+ +KL MF D+K S+D Q F H + ++ +++L G+W S + +
Sbjct: 487 ADYVNKLARMFQDIKVSEDLNQAFKEMHKNNKLALPADSVNIKILNAGAWSRSSEKVFVS 546
Query: 482 LPVEISALCEKFRSYYLGTHTGRRLSWQTNM--GFADLKATFGKGQKHELNVSTYQMCVL 539
LP E+ L + +Y H+GR+L W M G K G +++L V+T+Q+ VL
Sbjct: 547 LPTELEDLIPEVEEFYKKNHSGRKLHWHHLMSNGIITFKNEVG---QYDLEVTTFQLAVL 603
Query: 540 MLFNN--ADKLSYKEIEQATEIPAPDLKRCLQSLALVK--GRNVLRKEPM---SKDVGED 592
+N +K+S++ ++ ATE+P +L+R L SL R VL EP KD E
Sbjct: 604 FAWNQRPREKISFENLKLATELPDAELRRTLWSLVAFPKLKRQVLLYEPQVNSPKDFTEG 663
Query: 593 DAFSVNDKFS----SKLYK-VKIGTV-VAQKESEPEKQETRQRVEEDRKPQIEAAIVRIM 646
FSVN +FS +K+ K KI + Q +E ++E + + + R + ++IM
Sbjct: 664 TLFSVNQEFSLIKNAKVQKRGKINLIGRLQLTTERMREEENEGIVQLRILRTRKLYIQIM 723
Query: 647 KSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
K R+ + + L E+ + L+ FL +K++IE LIE ++ RD++D + Y+A
Sbjct: 724 KMRKKISNAQLQTELVEILKNMFLPQKKMIKEQIEWLIEHKYIRRDESDINTFIYMA 780
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 79,622,153
Number of Sequences: 164201
Number of extensions: 3341784
Number of successful extensions: 11099
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 10864
Number of HSP's gapped (non-prelim): 109
length of query: 703
length of database: 59,974,054
effective HSP length: 117
effective length of query: 586
effective length of database: 40,762,537
effective search space: 23886846682
effective search space used: 23886846682
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146568.7


