

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.8 - phase: 0 /pseudo
(430 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
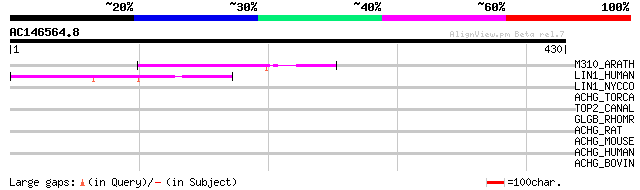
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 76 2e-13
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 49 2e-05
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 37 0.075
ACHG_TORCA (P02714) Acetylcholine receptor protein, gamma chain ... 33 1.1
TOP2_CANAL (P87078) DNA topoisomerase II (EC 5.99.1.3) 31 5.4
GLGB_RHOMR (Q93HU3) 1,4-alpha-glucan branching enzyme (EC 2.4.1.... 31 5.4
ACHG_RAT (P18916) Acetylcholine receptor protein, gamma chain pr... 30 9.2
ACHG_MOUSE (P04760) Acetylcholine receptor protein, gamma chain ... 30 9.2
ACHG_HUMAN (P07510) Acetylcholine receptor protein, gamma chain ... 30 9.2
ACHG_BOVIN (P13536) Acetylcholine receptor protein, gamma chain ... 30 9.2
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 75.9 bits (185), Expect = 2e-13
Identities = 45/160 (28%), Positives = 77/160 (48%), Gaps = 20/160 (12%)
Query: 100 AIPNFYLAFFKLPVKVWKRVVRIQREFLWGGVNGGKKVCWVKWATVCLPR-DKGGLGVRD 158
A+P + ++ F+L + K++ EF W +K+ WV W +C + D GGLG RD
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 159 MKLVNISLLAKWRWRLVQPEQPL*KDVLRCKYGSRINFL-----LDPIYNSWSSLASRWW 213
+ N +LLAK +R++ L +LR +Y + + P Y +W S+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSY-AWRSI----- 115
Query: 214 LDLMSLEGVVGANWFNREMVRKVGNGETTRFWLDHWVGNE 253
+ G +R ++R +G+G T+ WLD W+ +E
Sbjct: 116 --------IHGRELLSRGLLRTIGDGIHTKVWLDRWIMDE 147
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 49.3 bits (116), Expect = 2e-05
Identities = 41/176 (23%), Positives = 74/176 (41%), Gaps = 9/176 (5%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
L L+ F SG K+N KS R+ + + S +YLG+ + +
Sbjct: 717 LLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKYLGIQLTRDV 776
Query: 61 RRL--STWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVL--NAIPNFYLAFFKLPVKVW 116
+ L ++PLL+ +K+ N W N S GR+ ++ + I F KLP+ +
Sbjct: 777 KDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFF 836
Query: 117 KRVVRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRW 172
+ + +F+W +K + +T+ GG+ + D KL + + K W
Sbjct: 837 TELEKTTLKFIW-----NQKRAHIAKSTLSQKNKAGGITLPDFKLYYKATVTKTAW 887
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 37.4 bits (85), Expect = 0.075
Identities = 38/176 (21%), Positives = 73/176 (40%), Gaps = 9/176 (5%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
L +++ + SG K+N KS + + + +YLG+ + +
Sbjct: 717 LLEVIKEYSNVSGYKINTHKSVAFIYTNNNQAEKTVKDSIPFTVVPKKMKYLGVYLTKDV 776
Query: 61 RRL--STWEPLLDVLKKRLNSWGNKYVSLGGRV--VLLNAVLNAIPNFYLAFFKLPVKVW 116
+ L +E L + + +N W N S GR+ V ++ + AI NF K P+ +
Sbjct: 777 KDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYF 836
Query: 117 KRVVRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRW 172
K + +I F+W +K + + GG+ + D++L S++ K W
Sbjct: 837 KDLEKIILHFIW-----NQKKPQIAKTLLSNKNKAGGITLPDLRLYYKSIVIKTAW 887
>ACHG_TORCA (P02714) Acetylcholine receptor protein, gamma chain
precursor
Length = 506
Score = 33.5 bits (75), Expect = 1.1
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query: 246 LDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWN---LTWRRS 289
LDH + + L LT L S++ ++EA+ VW++ WN L+W S
Sbjct: 42 LDHII-DVTLKLTLTNLISLNEKEEALTTNVWIEIQWNDYRLSWNTS 87
>TOP2_CANAL (P87078) DNA topoisomerase II (EC 5.99.1.3)
Length = 1461
Score = 31.2 bits (69), Expect = 5.4
Identities = 22/83 (26%), Positives = 35/83 (41%), Gaps = 4/83 (4%)
Query: 59 NPRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKR 118
N RRL E L + + WG +G + ++ + I + + ++PVK W
Sbjct: 912 NIRRLMNGEELQE-MTPWYKGWGGDLEPMGPQKFKVSGRIEQIDSNTVEITEIPVKTWTN 970
Query: 119 VVRIQREFLWGGVNGGKKVCWVK 141
V +EFL G K W+K
Sbjct: 971 NV---KEFLLSGFGNEKTQPWIK 990
>GLGB_RHOMR (Q93HU3) 1,4-alpha-glucan branching enzyme (EC 2.4.1.18)
(Glycogen branching enzyme) (BE)
(1,4-alpha-D-glucan:1,4-alpha-D-glucan
6-glucosyl-transferase)
Length = 621
Score = 31.2 bits (69), Expect = 5.4
Identities = 29/122 (23%), Positives = 48/122 (38%), Gaps = 15/122 (12%)
Query: 252 NEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIHILLDELEGREV- 310
NE +YL FP +I+ + A G V L+ W G +H LD ++ +
Sbjct: 341 NETVYLHFPEAMTIAEESTAWPG---VSAPTYNNGLGFLYKWNMGWMHDTLDYIQRDPIY 397
Query: 311 -----SEPMDSWWWKLEEGGICSVSSSYSLHVK----LQMPPEPLENTK--AMVFGSVWK 359
E S W+ E + +S +H K +MP + + ++FG +W
Sbjct: 398 RKYHHDELTFSLWYAFSEHYVLPLSHDEVVHGKGSLWGKMPGDDWQKAANLRLLFGHMWG 457
Query: 360 SP 361
P
Sbjct: 458 HP 459
>ACHG_RAT (P18916) Acetylcholine receptor protein, gamma chain
precursor
Length = 519
Score = 30.4 bits (67), Expect = 9.2
Identities = 13/36 (36%), Positives = 21/36 (58%)
Query: 247 DHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDW 282
D V N +L LT L S++ ++EA+ VW++ W
Sbjct: 47 DSDVVNVSLKLTLTNLISLNEREEALTTNVWIEMQW 82
>ACHG_MOUSE (P04760) Acetylcholine receptor protein, gamma chain
precursor
Length = 519
Score = 30.4 bits (67), Expect = 9.2
Identities = 13/36 (36%), Positives = 21/36 (58%)
Query: 247 DHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDW 282
D V N +L LT L S++ ++EA+ VW++ W
Sbjct: 47 DSDVVNVSLKLTLTNLISLNEREEALTTNVWIEMQW 82
>ACHG_HUMAN (P07510) Acetylcholine receptor protein, gamma chain
precursor
Length = 517
Score = 30.4 bits (67), Expect = 9.2
Identities = 13/36 (36%), Positives = 21/36 (58%)
Query: 247 DHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDW 282
D V N +L LT L S++ ++EA+ VW++ W
Sbjct: 47 DSDVVNVSLKLTLTNLISLNEREEALTTNVWIEMQW 82
>ACHG_BOVIN (P13536) Acetylcholine receptor protein, gamma chain
precursor
Length = 519
Score = 30.4 bits (67), Expect = 9.2
Identities = 13/36 (36%), Positives = 21/36 (58%)
Query: 247 DHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDW 282
D V N +L LT L S++ ++EA+ VW++ W
Sbjct: 47 DSDVVNVSLKLTLTNLISLNEREEALTTNVWIEMQW 82
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.141 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,533,041
Number of Sequences: 164201
Number of extensions: 2486019
Number of successful extensions: 5480
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 5471
Number of HSP's gapped (non-prelim): 13
length of query: 430
length of database: 59,974,054
effective HSP length: 113
effective length of query: 317
effective length of database: 41,419,341
effective search space: 13129931097
effective search space used: 13129931097
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146564.8


