

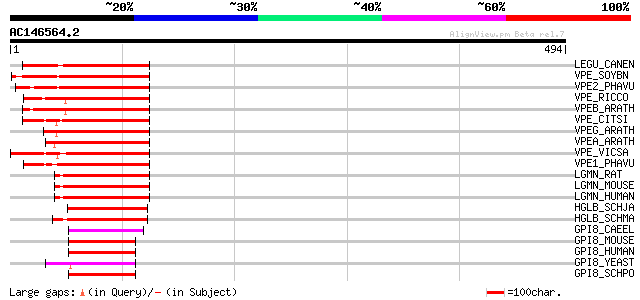
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.2 - phase: 0
(494 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LEGU_CANEN (P49046) Legumain precursor (EC 3.4.22.34) (Asparagin... 183 1e-45
VPE_SOYBN (P49045) Vacuolar processing enzyme precursor (EC 3.4.... 182 2e-45
VPE2_PHAVU (O24326) Vacuolar processing enzyme precursor (EC 3.4... 174 6e-43
VPE_RICCO (P49042) Vacuolar processing enzyme precursor (EC 3.4.... 154 4e-37
VPEB_ARATH (Q39044) Vacuolar processing enzyme, beta-isozyme pre... 154 5e-37
VPE_CITSI (P49043) Vacuolar processing enzyme precursor (EC 3.4.... 139 1e-32
VPEG_ARATH (Q39119) Vacuolar processing enzyme, gamma-isozyme pr... 138 4e-32
VPEA_ARATH (P49047) Vacuolar processing enzyme, alpha-isozyme pr... 138 4e-32
VPE_VICSA (P49044) Vacuolar processing enzyme precursor (EC 3.4.... 134 4e-31
VPE1_PHAVU (O24325) Vacuolar processing enzyme precursor (EC 3.4... 132 2e-30
LGMN_RAT (Q9R0J8) Legumain precursor (EC 3.4.22.34) (Asparaginyl... 115 3e-25
LGMN_MOUSE (O89017) Legumain precursor (EC 3.4.22.34) (Asparagin... 109 2e-23
LGMN_HUMAN (Q99538) Legumain precursor (EC 3.4.22.34) (Asparagin... 109 2e-23
HGLB_SCHJA (P42665) Hemoglobinase precursor (EC 3.4.22.34) (Anti... 97 1e-19
HGLB_SCHMA (P09841) Hemoglobinase precursor (EC 3.4.22.34) (Anti... 92 2e-18
GPI8_CAEEL (P49048) Potential GPI-anchor transamidase (EC 3.-.-.... 56 2e-07
GPI8_MOUSE (Q9CXY9) GPI-anchor transamidase precursor (EC 3.-.-.... 52 3e-06
GPI8_HUMAN (Q92643) GPI-anchor transamidase precursor (EC 3.-.-.... 52 3e-06
GPI8_YEAST (P49018) GPI-anchor transamidase precursor (EC 3.-.-.... 51 6e-06
GPI8_SCHPO (Q9USP5) GPI-anchor transamidase precursor (EC 3.-.-.... 46 3e-04
>LEGU_CANEN (P49046) Legumain precursor (EC 3.4.22.34) (Asparaginyl
endopeptidase)
Length = 475
Score = 183 bits (464), Expect = 1e-45
Identities = 89/113 (78%), Positives = 97/113 (85%), Gaps = 4/113 (3%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQAD 71
+L++L+L G AR N EWD VI+LP E VDD EVGTRWAVLVAGS+GYGNYRHQAD
Sbjct: 4 MLVMLSLHGTAARLNRREWDSVIQLPTEPVDD----EVGTRWAVLVAGSNGYGNYRHQAD 59
Query: 72 VCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
VCHAYQLLIKGGVKEENIVVFMYDDIA N +NPRPGVIINHPQGP+VY GVPK
Sbjct: 60 VCHAYQLLIKGGVKEENIVVFMYDDIAYNAMNPRPGVIINHPQGPDVYAGVPK 112
>VPE_SOYBN (P49045) Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE)
Length = 495
Score = 182 bits (461), Expect = 2e-45
Identities = 90/124 (72%), Positives = 104/124 (83%), Gaps = 6/124 (4%)
Query: 2 FSIVLSLWSWLLLLLT-LDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS 60
+S+VL W++++L + G ARPN EWD VI+LP E VD A+ DEVGTRWAVLVAGS
Sbjct: 14 YSVVL----WMMVVLVRVHGAAARPNRKEWDSVIKLPTEPVD-ADSDEVGTRWAVLVAGS 68
Query: 61 SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV 120
+GYGNYRHQADVCHAYQLLIKGG+KEENIVVFMYDDIA NELNPR GVIINHP+G ++Y
Sbjct: 69 NGYGNYRHQADVCHAYQLLIKGGLKEENIVVFMYDDIATNELNPRHGVIINHPEGEDLYA 128
Query: 121 GVPK 124
GVPK
Sbjct: 129 GVPK 132
>VPE2_PHAVU (O24326) Vacuolar processing enzyme precursor (EC
3.4.22.-) (Pv-VPE)
Length = 493
Score = 174 bits (440), Expect = 6e-43
Identities = 86/119 (72%), Positives = 99/119 (82%), Gaps = 4/119 (3%)
Query: 6 LSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGN 65
++ W W+L+++ + N E D VI+LP + VD AE DEVGTRWAVLVAGS+GYGN
Sbjct: 16 VAFWWWMLVMVMR---IQGTNGKEQDSVIKLPTQEVD-AESDEVGTRWAVLVAGSNGYGN 71
Query: 66 YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIA +ELNPRPGVIIN+PQGP+VY GVPK
Sbjct: 72 YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIATHELNPRPGVIINNPQGPDVYAGVPK 130
>VPE_RICCO (P49042) Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE)
Length = 497
Score = 154 bits (390), Expect = 4e-37
Identities = 80/116 (68%), Positives = 91/116 (77%), Gaps = 6/116 (5%)
Query: 13 LLLLTLDGLVA-RPNHLEWDPVIRLPGEVVDDAEVDE---VGTRWAVLVAGSSGYGNYRH 68
L L + GL+A R N E P I +P E + +VD+ +GTRWAVLVAGS G+GNYRH
Sbjct: 21 LSFLPIPGLLASRLNPFE--PGILMPTEEAEPVQVDDDDQLGTRWAVLVAGSMGFGNYRH 78
Query: 69 QADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
QADVCHAYQLL KGG+KEENI+VFMYDDIA NELNPRPGVIINHPQG +VY GVPK
Sbjct: 79 QADVCHAYQLLRKGGLKEENIIVFMYDDIAKNELNPRPGVIINHPQGEDVYAGVPK 134
>VPEB_ARATH (Q39044) Vacuolar processing enzyme, beta-isozyme
precursor (EC 3.4.22.-) (Beta-VPE)
Length = 486
Score = 154 bits (389), Expect = 5e-37
Identities = 79/115 (68%), Positives = 90/115 (77%), Gaps = 4/115 (3%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDE--VGTRWAVLVAGSSGYGNYRHQ 69
LLLLL L LV + ++P I +P E + A+ DE VGTRWAVLVAGSSGYGNYRHQ
Sbjct: 11 LLLLLVL--LVHAESRGRFEPKILMPTEEANPADQDEDGVGTRWAVLVAGSSGYGNYRHQ 68
Query: 70 ADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
ADVCHAYQ+L KGG+KEENIVV MYDDIAN+ LNPRPG +INHP G +VY GVPK
Sbjct: 69 ADVCHAYQILRKGGLKEENIVVLMYDDIANHPLNPRPGTLINHPDGDDVYAGVPK 123
>VPE_CITSI (P49043) Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE)
Length = 494
Score = 139 bits (351), Expect = 1e-32
Identities = 72/125 (57%), Positives = 89/125 (70%), Gaps = 14/125 (11%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEV------------VDDAEVDEVGTRWAVLVAG 59
+ LL+ L G+ + D +++LP E V+D + D VGTRWAVL+AG
Sbjct: 10 ITLLVALAGIADGSRDIAGD-ILKLPSEAYRFFHNGGGGAKVNDDD-DSVGTRWAVLLAG 67
Query: 60 SSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVY 119
S+G+ NYRHQAD+CHAYQLL KGG+K+ENI+VFMYDDIA NE NPRPGVIINHP G +VY
Sbjct: 68 SNGFWNYRHQADICHAYQLLRKGGLKDENIIVFMYDDIAFNEENPRPGVIINHPHGDDVY 127
Query: 120 VGVPK 124
GVPK
Sbjct: 128 KGVPK 132
>VPEG_ARATH (Q39119) Vacuolar processing enzyme, gamma-isozyme
precursor (EC 3.4.22.-) (Gamma-VPE)
Length = 490
Score = 138 bits (347), Expect = 4e-32
Identities = 68/100 (68%), Positives = 76/100 (76%), Gaps = 6/100 (6%)
Query: 31 DPVIRLPGEV------VDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGV 84
D VI+LP + ++ + GTRWAVLVAGSSGY NYRHQAD+CHAYQLL KGG+
Sbjct: 28 DDVIKLPSQASRFFRPAENDDDSNSGTRWAVLVAGSSGYWNYRHQADICHAYQLLRKGGL 87
Query: 85 KEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
KEENIVVFMYDDIANN NPRPG IIN P G +VY GVPK
Sbjct: 88 KEENIVVFMYDDIANNYENPRPGTIINSPHGKDVYQGVPK 127
>VPEA_ARATH (P49047) Vacuolar processing enzyme, alpha-isozyme
precursor (EC 3.4.22.-) (Alpha-VPE)
Length = 478
Score = 138 bits (347), Expect = 4e-32
Identities = 70/95 (73%), Positives = 75/95 (78%), Gaps = 3/95 (3%)
Query: 33 VIRLPG---EVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENI 89
VI+LP + E D+ T+WAVLVAGSSGY NYRHQADVCHAYQLL KGGVKEENI
Sbjct: 22 VIKLPSLASKFFRPTENDDDSTKWAVLVAGSSGYWNYRHQADVCHAYQLLKKGGVKEENI 81
Query: 90 VVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
VVFMYDDIA NE NPRPGVIIN P G +VY GVPK
Sbjct: 82 VVFMYDDIAKNEENPRPGVIINSPNGEDVYNGVPK 116
>VPE_VICSA (P49044) Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE) (Proteinase B)
Length = 493
Score = 134 bits (338), Expect = 4e-31
Identities = 75/133 (56%), Positives = 89/133 (66%), Gaps = 14/133 (10%)
Query: 1 MFSIVLSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVV---------DDAEVDEVGT 51
M S LS + +++T +V+ L D +RLP E DD E GT
Sbjct: 1 MGSSQLSTLLFFTIVVTFLTVVSSGRDLPGD-YLRLPSETSRFFREPKNDDDFE----GT 55
Query: 52 RWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIIN 111
RWA+L+AGS+GY NYRHQ+DVCHAYQLL KGG KEENI+VFMYDDIA+NE NPRPGVIIN
Sbjct: 56 RWAILLAGSNGYWNYRHQSDVCHAYQLLRKGGSKEENIIVFMYDDIASNEENPRPGVIIN 115
Query: 112 HPQGPNVYVGVPK 124
P G +VY GVPK
Sbjct: 116 KPDGDDVYAGVPK 128
>VPE1_PHAVU (O24325) Vacuolar processing enzyme precursor (EC
3.4.22.-) (VPE) (Legumain-like proteinase) (LLP)
Length = 484
Score = 132 bits (333), Expect = 2e-30
Identities = 73/113 (64%), Positives = 80/113 (70%), Gaps = 5/113 (4%)
Query: 13 LLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEV-GTRWAVLVAGSSGYGNYRHQAD 71
LLLL L LV+ L D +RLP D D V GTRWA+L AGSSGY NYRHQAD
Sbjct: 13 LLLLFLVALVSAGRDLVGD-FLRLPS---DSGNGDNVHGTRWAILFAGSSGYWNYRHQAD 68
Query: 72 VCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
+CHAYQLL KGG+K+ENI+VFMYDDIA N NPR GVIIN P G VY GVPK
Sbjct: 69 ICHAYQLLRKGGLKDENIIVFMYDDIAFNSENPRRGVIINSPNGDEVYKGVPK 121
>LGMN_RAT (Q9R0J8) Legumain precursor (EC 3.4.22.34) (Asparaginyl
endopeptidase) (Protease, cysteine 1)
Length = 435
Score = 115 bits (287), Expect = 3e-25
Identities = 52/84 (61%), Positives = 64/84 (75%), Gaps = 2/84 (2%)
Query: 41 VDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANN 100
VDD E + G W V+VAGS+G+ NYRHQAD CHAYQ++ + G+ +E I+V MYDDIANN
Sbjct: 22 VDDPE--DGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIIVMMYDDIANN 79
Query: 101 ELNPRPGVIINHPQGPNVYVGVPK 124
E NP PGV+IN P G +VY GVPK
Sbjct: 80 EENPTPGVVINRPNGTDVYKGVPK 103
>LGMN_MOUSE (O89017) Legumain precursor (EC 3.4.22.34) (Asparaginyl
endopeptidase) (Protease, cysteine 1)
Length = 435
Score = 109 bits (272), Expect = 2e-23
Identities = 50/84 (59%), Positives = 63/84 (74%), Gaps = 2/84 (2%)
Query: 41 VDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANN 100
VDD E + G W V+VAGS+G+ NYRHQAD CHAYQ++ + G+ +E I+V MYDDIAN+
Sbjct: 22 VDDPE--DGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIIVMMYDDIANS 79
Query: 101 ELNPRPGVIINHPQGPNVYVGVPK 124
E NP PGV+IN P G +VY GV K
Sbjct: 80 EENPTPGVVINRPNGTDVYKGVLK 103
>LGMN_HUMAN (Q99538) Legumain precursor (EC 3.4.22.34) (Asparaginyl
endopeptidase) (Protease, cysteine 1)
Length = 433
Score = 109 bits (272), Expect = 2e-23
Identities = 49/84 (58%), Positives = 63/84 (74%), Gaps = 2/84 (2%)
Query: 41 VDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANN 100
+DD E + G W V+VAGS+G+ NYRHQAD CHAYQ++ + G+ +E IVV MYDDIA +
Sbjct: 20 IDDPE--DGGKHWVVIVAGSNGWYNYRHQADACHAYQIIHRNGIPDEQIVVMMYDDIAYS 77
Query: 101 ELNPRPGVIINHPQGPNVYVGVPK 124
E NP PG++IN P G +VY GVPK
Sbjct: 78 EDNPTPGIVINRPNGTDVYQGVPK 101
>HGLB_SCHJA (P42665) Hemoglobinase precursor (EC 3.4.22.34) (Antigen
Sj32)
Length = 423
Score = 96.7 bits (239), Expect = 1e-19
Identities = 44/71 (61%), Positives = 55/71 (76%)
Query: 52 RWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIIN 111
+WAVLVAGS+G+ NYRHQADVCHAY +L+ GVK E+I+ FMYDDIA+N+ NP PG I N
Sbjct: 31 KWAVLVAGSNGFENYRHQADVCHAYHVLLSKGVKPEHIITFMYDDIAHNKENPFPGKIFN 90
Query: 112 HPQGPNVYVGV 122
+ + Y GV
Sbjct: 91 DYRHKDYYKGV 101
>HGLB_SCHMA (P09841) Hemoglobinase precursor (EC 3.4.22.34) (Antigen
SM32)
Length = 429
Score = 92.4 bits (228), Expect = 2e-18
Identities = 46/84 (54%), Positives = 56/84 (65%), Gaps = 2/84 (2%)
Query: 39 EVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIA 98
EV D+ D +WAVLVAGS+GY NYRHQADVCHAY +L G+K E+I+ MYDDIA
Sbjct: 26 EVSDETVSDN--NKWAVLVAGSNGYPNYRHQADVCHAYHVLRSKGIKPEHIITMMYDDIA 83
Query: 99 NNELNPRPGVIINHPQGPNVYVGV 122
N +NP PG + N + Y GV
Sbjct: 84 YNLMNPFPGKLFNDYNHKDWYEGV 107
>GPI8_CAEEL (P49048) Potential GPI-anchor transamidase (EC 3.-.-.-)
(GPI transamidase)
Length = 322
Score = 55.8 bits (133), Expect = 2e-07
Identities = 25/67 (37%), Positives = 39/67 (57%)
Query: 53 WAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINH 112
WAVLV S + NYRH ++V Y + + G+ + NI++ + +D+ N NPRPG +
Sbjct: 42 WAVLVCTSKFWFNYRHVSNVLALYHSIKRLGIPDSNIIMMLAEDVPCNSRNPRPGTVYAA 101
Query: 113 PQGPNVY 119
G N+Y
Sbjct: 102 RAGTNLY 108
>GPI8_MOUSE (Q9CXY9) GPI-anchor transamidase precursor (EC 3.-.-.-)
(GPI transamidase) (Phosphatidylinositol-glycan
biosynthesis, class K protein) (PIG-K)
Length = 395
Score = 52.4 bits (124), Expect = 3e-06
Identities = 23/60 (38%), Positives = 37/60 (61%)
Query: 53 WAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINH 112
WAVLV S + NYRH A+ Y+ + + G+ + +IV+ + DD+A N NP+P + +H
Sbjct: 46 WAVLVCTSRFWFNYRHVANTLSVYRSVKRLGIPDSHIVLMLADDMACNARNPKPATVFSH 105
>GPI8_HUMAN (Q92643) GPI-anchor transamidase precursor (EC 3.-.-.-)
(GPI transamidase) (Phosphatidylinositol-glycan
biosynthesis, class K protein) (PIG-K) (hGPI8)
Length = 395
Score = 52.4 bits (124), Expect = 3e-06
Identities = 23/60 (38%), Positives = 37/60 (61%)
Query: 53 WAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINH 112
WAVLV S + NYRH A+ Y+ + + G+ + +IV+ + DD+A N NP+P + +H
Sbjct: 46 WAVLVCTSRFWFNYRHVANTLSVYRSVKRLGIPDSHIVLMLADDMACNPRNPKPATVFSH 105
>GPI8_YEAST (P49018) GPI-anchor transamidase precursor (EC 3.-.-.-)
(GPI transamidase)
Length = 411
Score = 51.2 bits (121), Expect = 6e-06
Identities = 28/83 (33%), Positives = 46/83 (54%), Gaps = 3/83 (3%)
Query: 33 VIRLPGEVVDDAEVDEVGTR---WAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENI 89
++ L G DA + + T WAVLV+ S + NYRH A+V Y+ + + G+ + I
Sbjct: 16 LLPLSGANNTDAAHEVIATNTNNWAVLVSTSRFWFNYRHMANVLSMYRTVKRLGIPDSQI 75
Query: 90 VVFMYDDIANNELNPRPGVIINH 112
++ + DD+A N N PG + N+
Sbjct: 76 ILMLSDDVACNSRNLFPGSVFNN 98
>GPI8_SCHPO (Q9USP5) GPI-anchor transamidase precursor (EC 3.-.-.-)
(GPI transamidase)
Length = 380
Score = 45.8 bits (107), Expect = 3e-04
Identities = 21/60 (35%), Positives = 36/60 (60%)
Query: 53 WAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINH 112
WAVL++ S + NYRH A+V Y+ + + G+ + I++ + DD A N N PG + ++
Sbjct: 27 WAVLISTSRFWFNYRHTANVLGIYRSVKRLGIPDSQIILMIADDYACNSRNLFPGTVFDN 86
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.348 0.155 0.614
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,168,617
Number of Sequences: 164201
Number of extensions: 2399309
Number of successful extensions: 6560
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 6536
Number of HSP's gapped (non-prelim): 22
length of query: 494
length of database: 59,974,054
effective HSP length: 114
effective length of query: 380
effective length of database: 41,255,140
effective search space: 15676953200
effective search space used: 15676953200
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146564.2


