

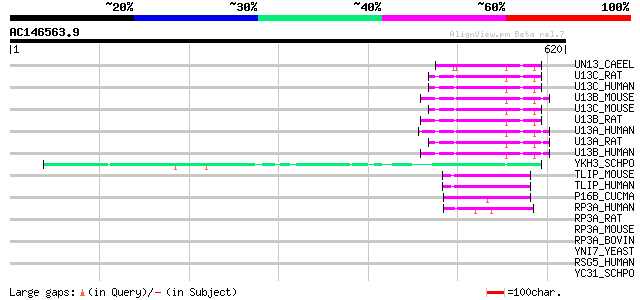
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.9 + phase: 0
(620 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UN13_CAEEL (P27715) Phorbol ester/diacylglycerol-binding protein... 61 1e-08
U13C_RAT (Q62770) Unc-13 homolog C (Munc13-3) 58 8e-08
U13C_HUMAN (Q8NB66) Unc-13 homolog C (Munc13-3) (Fragment) 58 8e-08
U13B_MOUSE (Q9Z1N9) Unc-13 homolog B (Munc13-2) (munc13) 58 8e-08
U13C_MOUSE (Q8K0T7) Unc-13 homolog C (Munc13-3) (Fragment) 57 1e-07
U13B_RAT (Q62769) Unc-13 homolog B (Munc13-2) 57 1e-07
U13A_HUMAN (Q9UPW8) Unc-13 homolog A (Munc13-1) (Fragment) 57 1e-07
U13A_RAT (Q62768) Unc-13 homolog A (Munc13-1) 57 2e-07
U13B_HUMAN (O14795) Unc-13 homolog B (Munc13-2) (munc13) 55 4e-07
YKH3_SCHPO (Q9UT00) Hypothetical protein PYUK71.03c in chromosome I 55 5e-07
TLIP_MOUSE (Q9QZ06) Toll-interacting protein 51 8e-06
TLIP_HUMAN (Q9H0E2) Toll-interacting protein 50 2e-05
P16B_CUCMA (Q9ZT46) 16 kDa phloem protein 2 47 1e-04
RP3A_HUMAN (Q9Y2J0) Rabphilin-3A (Exophilin 1) 45 7e-04
RP3A_RAT (P47709) Rabphilin-3A (Exophilin 1) 44 0.001
RP3A_MOUSE (P47708) Rabphilin-3A (Exophilin 1) 44 0.001
RP3A_BOVIN (Q06846) Rabphilin-3A (Exophilin 1) 44 0.001
YNI7_YEAST (P48231) Hypothetical 132.5 kDa protein in TOP2-MKT1 ... 42 0.004
RSG5_HUMAN (O43374) Ras GTPase-activating protein 4 (RasGAP-acti... 42 0.004
YC31_SCHPO (O14065) Hypothetical protein C962.01 in chromosome III 42 0.005
>UN13_CAEEL (P27715) Phorbol ester/diacylglycerol-binding protein
unc-13 (Uncoordinated protein 13)
Length = 2155
Score = 60.8 bits (146), Expect = 1e-08
Identities = 48/151 (31%), Positives = 70/151 (45%), Gaps = 34/151 (22%)
Query: 476 NIQLRTGKKLKITVVEGKD----------LAA----AKEKTGKFDPYIKLQYGKVMQKTK 521
NIQ T K +K +++EG L A AK+KTGK DPY+ Q GK ++T+
Sbjct: 1135 NIQKETLKTVKASILEGSSKWSAKITLTVLCAQGLIAKDKTGKSDPYVTAQVGKTKRRTR 1194
Query: 522 TSHTP-NPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVNLE 568
T H NPVWN+ F+ + +K++V+ E+ D+ +G + +
Sbjct: 1195 TIHQELNPVWNEKFHFECHNSTDRIKVRVWDEDNDLKSKLRQKLTRESDDFLGQTVIEVR 1254
Query: 569 GLVDGSVRDVWIPLER-----VRSGEIRLKI 594
L DVW LE+ SG IRL I
Sbjct: 1255 TL--SGEMDVWYNLEKRTDKSAVSGAIRLHI 1283
>U13C_RAT (Q62770) Unc-13 homolog C (Munc13-3)
Length = 2204
Score = 57.8 bits (138), Expect = 8e-08
Identities = 48/145 (33%), Positives = 71/145 (48%), Gaps = 25/145 (17%)
Query: 468 QQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TP 526
Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++TKT
Sbjct: 1199 QSVLDGTS----KWSAKITITVVSAQGLQA-KDKTGSSDPYVTVQVGKNKRRTKTIFGNL 1253
Query: 527 NPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVNLEGLVDGS 574
NPVW++ F+ + +K++V+ E+ D+ +G V + L
Sbjct: 1254 NPVWDEKFYFECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTL--SG 1311
Query: 575 VRDVWIPLER-----VRSGEIRLKI 594
DVW LE+ SG IRLKI
Sbjct: 1312 EMDVWYNLEKRTDKSAVSGAIRLKI 1336
>U13C_HUMAN (Q8NB66) Unc-13 homolog C (Munc13-3) (Fragment)
Length = 1190
Score = 57.8 bits (138), Expect = 8e-08
Identities = 48/145 (33%), Positives = 71/145 (48%), Gaps = 25/145 (17%)
Query: 468 QQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TP 526
Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++TKT
Sbjct: 185 QSVLDGTS----KWSAKITITVVSAQGLQA-KDKTGSSDPYVTVQVGKNKRRTKTIFGNL 239
Query: 527 NPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVNLEGLVDGS 574
NPVW++ F+ + +K++V+ E+ D+ +G V + L
Sbjct: 240 NPVWDEKFYFECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTL--SG 297
Query: 575 VRDVWIPLER-----VRSGEIRLKI 594
DVW LE+ SG IRLKI
Sbjct: 298 EMDVWYNLEKRTDKSAVSGAIRLKI 322
>U13B_MOUSE (Q9Z1N9) Unc-13 homolog B (Munc13-2) (munc13)
Length = 1591
Score = 57.8 bits (138), Expect = 8e-08
Identities = 47/162 (29%), Positives = 79/162 (48%), Gaps = 26/162 (16%)
Query: 460 LNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
+ ++ Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++
Sbjct: 581 VQQMKTVKQSVLDGTS----KWSAKITITVVCAQGLQA-KDKTGSSDPYVTVQVGKTKKR 635
Query: 520 TKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVN 566
TKT NPVW + F+ + +K++V+ E+ D+ +G +
Sbjct: 636 TKTIFGNLNPVWEEKFHFECHNSSDRIKVRVWDEDDDIKSRVKQRLKRESDDFLGQTIIE 695
Query: 567 LEGLVDGSVRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
+ L DVW LE+ SG IRL+I ++++ +E
Sbjct: 696 VRTL--SGEMDVWYNLEKRTDKSAVSGAIRLQI-SVEIKGEE 734
>U13C_MOUSE (Q8K0T7) Unc-13 homolog C (Munc13-3) (Fragment)
Length = 1430
Score = 57.4 bits (137), Expect = 1e-07
Identities = 48/145 (33%), Positives = 71/145 (48%), Gaps = 25/145 (17%)
Query: 468 QQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TP 526
Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++TKT
Sbjct: 1205 QSVLDGTS----KWSAKITITVVSAQGLQA-KDKTGSSDPYVTVQVGKNKRRTKTIFGNL 1259
Query: 527 NPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVNLEGLVDGS 574
NPVW++ F+ + +K++V+ E+ D+ +G V + L
Sbjct: 1260 NPVWDEKFFFECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTL--SG 1317
Query: 575 VRDVWIPLER-----VRSGEIRLKI 594
DVW LE+ SG IRLKI
Sbjct: 1318 EMDVWYNLEKRTDKSAVSGAIRLKI 1342
>U13B_RAT (Q62769) Unc-13 homolog B (Munc13-2)
Length = 1622
Score = 57.4 bits (137), Expect = 1e-07
Identities = 46/153 (30%), Positives = 73/153 (47%), Gaps = 25/153 (16%)
Query: 460 LNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
+ ++ Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++
Sbjct: 594 VQQMKTVKQSVLDGTS----KWSAKITITVVCAQGLQA-KDKTGSSDPYVTVQVGKTKKR 648
Query: 520 TKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVN 566
TKT NPVW + F+ + +K++V+ E+ D+ +G +
Sbjct: 649 TKTIFGNLNPVWEEKFHFECHNSSDRIKVRVWDEDDDIKSRVKQRLKRESDDFLGQTIIE 708
Query: 567 LEGLVDGSVRDVWIPLER-----VRSGEIRLKI 594
+ L DVW LE+ SG IRL+I
Sbjct: 709 VRTL--SGEMDVWYNLEKRTDKSAVSGAIRLQI 739
>U13A_HUMAN (Q9UPW8) Unc-13 homolog A (Munc13-1) (Fragment)
Length = 1157
Score = 57.4 bits (137), Expect = 1e-07
Identities = 48/165 (29%), Positives = 81/165 (49%), Gaps = 27/165 (16%)
Query: 457 THSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKV 516
TH+ ++ Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK
Sbjct: 109 THT-QQMKAVKQSVLDGTS----KWSAKISITVVCAQGLQA-KDKTGSSDPYVTVQVGKT 162
Query: 517 MQKTKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSA 563
++TKT + NPVW + F+ + +K++V+ E+ D+ +G
Sbjct: 163 KKRTKTIYGNLNPVWEENFHFECHNSSDRIKVRVWDEDDDIKSRVKQRFKRESDDFLGQT 222
Query: 564 QVNLEGLVDGSVRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
+ + L DVW L++ SG IRL I ++++ +E
Sbjct: 223 IIEVRTL--SGEMDVWYNLDKRTDKSAVSGAIRLHI-SVEIKGEE 264
>U13A_RAT (Q62768) Unc-13 homolog A (Munc13-1)
Length = 1735
Score = 56.6 bits (135), Expect = 2e-07
Identities = 46/154 (29%), Positives = 75/154 (47%), Gaps = 26/154 (16%)
Query: 468 QQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TP 526
Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q GK ++TKT +
Sbjct: 678 QSVLDGTS----KWSAKISITVVCAQGLQA-KDKTGSSDPYVTVQVGKTKKRTKTIYGNL 732
Query: 527 NPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVNLEGLVDGS 574
NPVW + F+ + +K++V E+ D+ +G + + L
Sbjct: 733 NPVWEENFHFECHNSSDRIKVRVLDEDDDIKSRVKQRFKRESDDFLGQTIIEVRTL--SG 790
Query: 575 VRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
DVW L++ SG IRL I ++++ +E
Sbjct: 791 EMDVWYNLDKRTDKSAVSGAIRLHI-SVEIKGEE 823
>U13B_HUMAN (O14795) Unc-13 homolog B (Munc13-2) (munc13)
Length = 1591
Score = 55.5 bits (132), Expect = 4e-07
Identities = 46/162 (28%), Positives = 78/162 (47%), Gaps = 26/162 (16%)
Query: 460 LNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
+ ++ Q L+G+S + K+ ITVV + L A K+KTG DPY+ +Q K ++
Sbjct: 581 VQQMKTVKQSVLDGTS----KWSAKITITVVCAQGLQA-KDKTGSSDPYVTVQVSKTKKR 635
Query: 520 TKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVN 566
TKT NPVW + F+ + +K++V+ E+ D+ +G +
Sbjct: 636 TKTIFGNLNPVWEEKFHFECHNSSDRIKVRVWDEDDDIKSRVKQRLKRESDDFLGQTIIE 695
Query: 567 LEGLVDGSVRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
+ L DVW LE+ SG IRL+I ++++ +E
Sbjct: 696 VRTL--SGEMDVWYNLEKRTDKSAVSGAIRLQI-SVEIKGEE 734
>YKH3_SCHPO (Q9UT00) Hypothetical protein PYUK71.03c in chromosome I
Length = 1225
Score = 55.1 bits (131), Expect = 5e-07
Identities = 111/583 (19%), Positives = 222/583 (38%), Gaps = 84/583 (14%)
Query: 38 ILIACAVEKWVFSFSTW--VPLALAVWATIQYGRYQRKLLVEDLDKKWKRIILNNSPITP 95
+++ AV ++F + + + L + + IQY R + + + + R +
Sbjct: 158 LVLYTAVMSFLFGYLRFGFLSLFIIMAVCIQYYRICDRRVKVNFKDDYTRYLSTRKLEND 217
Query: 96 LEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSL 155
E WLN L + W F P LS R++ I + L P F++ + L EF+LG+ P +
Sbjct: 218 SETVTWLNTFLQQFW-YIFEPSLSERITEITDQILSENVPSFIDSMALSEFTLGTKSPRM 276
Query: 156 ALQGMRWSTIGDQRVMQLGFDWDTHEMS--------------ILLLAKLAKPLMGTAR-I 200
T D +M L + +++S I L K+ K + +
Sbjct: 277 GFIRSYPKTEEDTVMMDLRLAFSPNDISDLTGREIAACIKPKIALDLKIGKSIASAKMPV 336
Query: 201 VINSLHIKGDL-IFTPILD----GKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVS 255
+I L G+L + ++D K + +F P + G PG++
Sbjct: 337 LIEDLSFTGNLRVKVKLIDKYPYAKTVGLTFTEKPVFSYILKPLGGDKFGFDIGNIPGLT 396
Query: 256 SWLEKLFTDTLVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQ 315
+++ + +TL M P +L +++ G + L + FK +
Sbjct: 397 TFITEQIHNTLGPMMYSPN-------VYELDIESMMGAAGLN----TALGAVEFKLRK-- 443
Query: 316 QSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGS-TPRWDAPFNMVLHDNTGTLR 374
+G + + D + ++ + + ++ V + +P ++ F VL+ + L
Sbjct: 444 ----GDGFKDGLGGAVDPYVVIKNSADRVIGKSKVAHNTGSPVFNETFYSVLNSFSENLN 499
Query: 375 FNLYECIPNNVKCD-YLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVP 433
+Y+ N+++ D LGS + L +E V D+ + E+ +
Sbjct: 500 LEVYDF--NDIRSDKLLGSAVLPLATLE-----AMPVTNDAFV----------ELTLKGK 542
Query: 434 FEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGK 493
G + ++K VV + +G ++ L+ TV + K
Sbjct: 543 TVGRLNYDMKFHAVVPD---------------------SGEEITKVDGPGVLQFTVHQCK 581
Query: 494 DLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTI-EFDEVGGGEYLKLKVFTE 552
+L+ K + + +V K NP W ++ G L +++FTE
Sbjct: 582 ELSNDPSKRPTAYAKLIINNKEVYTTRKIKKNNNPSWEESFGTLLPEGKNATLGVQIFTE 641
Query: 553 ELFGDENIGSAQVNLEGLVDGSVRD-VWIPLERVRSGEIRLKI 594
E + G+A V+L+ L + +W PL+ SG +R+ +
Sbjct: 642 E--SEHPFGTANVSLQDLFAATKTGLLWFPLQHAPSGRVRMSV 682
>TLIP_MOUSE (Q9QZ06) Toll-interacting protein
Length = 274
Score = 51.2 bits (121), Expect = 8e-06
Identities = 34/103 (33%), Positives = 50/103 (48%), Gaps = 6/103 (5%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHT--PNPVWNQTIEFDEVGG 541
+L ITVV+ K A + DPY +L+ G + +T T+H NP WN+ I+ G
Sbjct: 54 RLSITVVQAK--LAKNYGMTRMDPYCRLRLGYAVYETPTAHNGAKNPRWNKVIQCTVPPG 111
Query: 542 GEYLKLKVFTEELFG-DENIGSAQVNL-EGLVDGSVRDVWIPL 582
+ L++F E F D+ I + + E L G V D W L
Sbjct: 112 VDSFYLEIFDERAFSMDDRIAWTHITIPESLKQGQVEDEWYSL 154
>TLIP_HUMAN (Q9H0E2) Toll-interacting protein
Length = 274
Score = 50.1 bits (118), Expect = 2e-05
Identities = 34/103 (33%), Positives = 49/103 (47%), Gaps = 6/103 (5%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHT--PNPVWNQTIEFDEVGG 541
+L ITVV+ K A + DPY +L+ G + +T T+H NP WN+ I G
Sbjct: 54 RLNITVVQAK--LAKNYGMTRMDPYCRLRLGYAVYETPTAHNGAKNPRWNKVIHCTVPPG 111
Query: 542 GEYLKLKVFTEELFG-DENIGSAQVNL-EGLVDGSVRDVWIPL 582
+ L++F E F D+ I + + E L G V D W L
Sbjct: 112 VDSFYLEIFDERAFSMDDRIAWTHITIPESLRQGKVEDKWYSL 154
>P16B_CUCMA (Q9ZT46) 16 kDa phloem protein 2
Length = 137
Score = 47.4 bits (111), Expect = 1e-04
Identities = 27/105 (25%), Positives = 53/105 (49%), Gaps = 7/105 (6%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYG--KVMQKTKTSHTPNPVWNQ----TIEFDE 538
+++ ++ GK L A DPY ++ + + M K + P+P+WN+ +E+
Sbjct: 5 MEVHLISGKGLQAHDPLNKPIDPYAEINFKGQERMSKVAKNAGPDPIWNEKFKFLVEYPG 64
Query: 539 VGGGEYLKLKVFTEE-LFGDENIGSAQVNLEGLVDGSVRDVWIPL 582
GG ++ KV + + GD+ IG +++++ L+ VR W L
Sbjct: 65 SGGDFHILFKVMDHDAIDGDDYIGDVKIDVQNLLAEGVRKGWSEL 109
>RP3A_HUMAN (Q9Y2J0) Rabphilin-3A (Exophilin 1)
Length = 694
Score = 44.7 bits (104), Expect = 7e-04
Identities = 37/112 (33%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK-----TKT-SHTPNPVWNQTIEF-- 536
L+ T+++ K L + G DPY+KL K TKT +T NP+WN+T+ +
Sbjct: 409 LQCTIIKAKGLKPM-DSNGLADPYVKLHLLPGASKSNKLRTKTLRNTRNPIWNETLVYHG 467
Query: 537 --DEVGGGEYLKLKVFTEELFG-DENIGSAQVNLEGLVDGSVRDVWIPLERV 585
DE + L++ V E+ FG +E IG + +L+ L ++ I LERV
Sbjct: 468 ITDEDMQRKTLRISVCDEDKFGHNEFIGETRFSLKKLKPNQRKNFNICLERV 519
>RP3A_RAT (P47709) Rabphilin-3A (Exophilin 1)
Length = 684
Score = 44.3 bits (103), Expect = 0.001
Identities = 37/112 (33%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK-----TKT-SHTPNPVWNQTIEF-- 536
L+ T++ K L + G DPY+KL K TKT +T NPVWN+T+++
Sbjct: 399 LQCTIIRAKGLKPM-DSNGLADPYVKLHLLPGASKSNKLRTKTLRNTRNPVWNETLQYHG 457
Query: 537 --DEVGGGEYLKLKVFTEELFG-DENIGSAQVNLEGLVDGSVRDVWIPLERV 585
+E + L++ V E+ FG +E IG + +L+ L ++ I LERV
Sbjct: 458 ITEEDMQRKTLRISVCDEDKFGHNEFIGETRFSLKKLKANQRKNFNICLERV 509
>RP3A_MOUSE (P47708) Rabphilin-3A (Exophilin 1)
Length = 681
Score = 44.3 bits (103), Expect = 0.001
Identities = 37/112 (33%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK-----TKT-SHTPNPVWNQTIEF-- 536
L+ T++ K L + G DPY+KL K TKT +T NPVWN+T+++
Sbjct: 396 LQCTIIRAKGLKPM-DSNGLADPYVKLHLLPGASKSNKLRTKTLRNTRNPVWNETLQYHG 454
Query: 537 --DEVGGGEYLKLKVFTEELFG-DENIGSAQVNLEGLVDGSVRDVWIPLERV 585
+E + L++ V E+ FG +E IG + +L+ L ++ I LERV
Sbjct: 455 ITEEDMQRKTLRISVCDEDKFGHNEFIGETRFSLKKLKANQRKNFNICLERV 506
>RP3A_BOVIN (Q06846) Rabphilin-3A (Exophilin 1)
Length = 704
Score = 43.9 bits (102), Expect = 0.001
Identities = 37/112 (33%), Positives = 57/112 (50%), Gaps = 12/112 (10%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK-----TKT-SHTPNPVWNQTIEF-- 536
L T+++ K L + G DPY+KL K TKT +T NP+WN+T+ +
Sbjct: 419 LHCTIIKAKGLKPM-DSNGLADPYVKLHLLPGASKSNKLRTKTLRNTRNPIWNETLVYHG 477
Query: 537 --DEVGGGEYLKLKVFTEELFG-DENIGSAQVNLEGLVDGSVRDVWIPLERV 585
DE + L++ V E+ FG +E IG + +L+ L ++ I LERV
Sbjct: 478 ITDEDMQRKTLRISVCDEDKFGHNEFIGETRFSLKKLKPNQRKNFNICLERV 529
>YNI7_YEAST (P48231) Hypothetical 132.5 kDa protein in TOP2-MKT1
intergenic region
Length = 1178
Score = 42.4 bits (98), Expect = 0.004
Identities = 39/157 (24%), Positives = 69/157 (43%), Gaps = 10/157 (6%)
Query: 35 IPVILIA--CAVEKWVFSFSTWVPLALAVWATIQYGRYQRKLL--VEDLDKKWKRIILNN 90
+ +++IA C+ F FS L + + + Y +K + DL +K + +
Sbjct: 103 VAILIIAGLCSFVLGYFKFSLASVLIVMLTTGMLYRTSSKKYRESLRDLAQKEQTV---E 159
Query: 91 SPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLK--LRKPRFLERVELQEFSL 148
+ E EWLN L + WP P +S ++ L + P+F++ + L +F+L
Sbjct: 160 KITSDYESVEWLNTFLDKYWP-IIEPSVSQQIVDGTNTALSENVAIPKFIKAIWLDQFTL 218
Query: 149 GSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSIL 185
G PP + +T D VM + + H+MS L
Sbjct: 219 GVKPPRIDAIKTFQNTKSDVVVMDVCLSFTPHDMSDL 255
Score = 35.8 bits (81), Expect = 0.34
Identities = 28/107 (26%), Positives = 50/107 (46%), Gaps = 25/107 (23%)
Query: 480 RTGKKLKITVVEGKDLAAAKEKTGKF----DPYIKLQY-GKVMQKTKT-SHTPNPVWNQT 533
+TG + + ++ K+ ++ G DPY+ + GK++ KTK ++ NPVWN++
Sbjct: 372 KTGLPIGVLEIKVKNAHGLRKLVGMIKKTVDPYLTFELSGKIVGKTKVFKNSANPVWNES 431
Query: 534 IEFDEVGGGEYLKLKVFTE----------ELFGDENIGSAQVNLEGL 570
I Y+ L+ FT+ E D+ +G+ NL L
Sbjct: 432 I---------YILLQSFTDPLTIAVYDKRETLSDKKMGTVIFNLNKL 469
Score = 33.5 bits (75), Expect = 1.7
Identities = 19/74 (25%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 500 EKTGKFDPYIKLQYGKVMQ--KTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEELFGD 557
+ GK PY K+ V + + T NP+WNQ+I + + + F + GD
Sbjct: 666 DSLGKISPYAKVSVNGVARGRTNERIETLNPIWNQSIYVSVTSPLQKVSIDCFGIDTNGD 725
Query: 558 E-NIGSAQVNLEGL 570
+ N+GS + + +
Sbjct: 726 DHNLGSLNIQTQNI 739
>RSG5_HUMAN (O43374) Ras GTPase-activating protein 4
(RasGAP-activating-like protein 2) (Calcium-promoted Ras
inactivator)
Length = 803
Score = 42.4 bits (98), Expect = 0.004
Identities = 26/103 (25%), Positives = 58/103 (56%), Gaps = 4/103 (3%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQY-GKVMQKTKTSHTPNPVWNQTIEFD-EVGG 541
+L+ +V+E +DLA K++ G DP+++++Y G+ + + + P WN+T EF+ + G
Sbjct: 134 RLRCSVLEARDLAP-KDRNGTSDPFVRVRYKGRTRETSIVKKSCYPRWNETFEFELQEGA 192
Query: 542 GEYLKLKVFTEELFG-DENIGSAQVNLEGLVDGSVRDVWIPLE 583
E L ++ + +L ++ +G ++++ L + W L+
Sbjct: 193 MEALCVEAWDWDLVSRNDFLGKVVIDVQRLRVVQQEEGWFRLQ 235
>YC31_SCHPO (O14065) Hypothetical protein C962.01 in chromosome III
Length = 1429
Score = 42.0 bits (97), Expect = 0.005
Identities = 35/128 (27%), Positives = 56/128 (43%), Gaps = 19/128 (14%)
Query: 38 ILIACAVEKWVFS---FSTWVPLALAVWATIQYG-------RYQRKLLVEDLDKKWKRII 87
IL+A ++ W+ S F ++ + + TI YG R R+ +V++L KK
Sbjct: 207 ILLAVSILSWIASKLWFRFFILFFIIITGTIVYGSCMISVRRNIREEVVQELSKK----- 261
Query: 88 LNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFS 147
N + E W N LL W P++S +S VE + P F++ F+
Sbjct: 262 --NGDVD-YETMSWFNTLLQRFWM-LNEPEISKSVSTSVEQSIAEYLPSFIKEAAFSTFT 317
Query: 148 LGSCPPSL 155
LGS P +
Sbjct: 318 LGSKAPRI 325
Score = 41.2 bits (95), Expect = 0.008
Identities = 24/96 (25%), Positives = 50/96 (52%), Gaps = 5/96 (5%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPN---PVWNQTIEFDEVGG 541
++++V++ DL + T K DPY ++ G + +T +TPN P+WN+ + +
Sbjct: 759 MRLSVIKANDLVNVELPTRKSDPYARVIVGNSVV-ARTVYTPNNLNPIWNEILYVPIMAD 817
Query: 542 GEYLKLKVFT-EELFGDENIGSAQVNLEGLVDGSVR 576
+ + L+ EE D ++G A +N++ + + R
Sbjct: 818 TKTIDLEAMDYEESGNDRSLGYASINVQKYIRNAKR 853
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 74,161,685
Number of Sequences: 164201
Number of extensions: 3209551
Number of successful extensions: 8116
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 8046
Number of HSP's gapped (non-prelim): 116
length of query: 620
length of database: 59,974,054
effective HSP length: 116
effective length of query: 504
effective length of database: 40,926,738
effective search space: 20627075952
effective search space used: 20627075952
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146563.9


