

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.12 - phase: 0
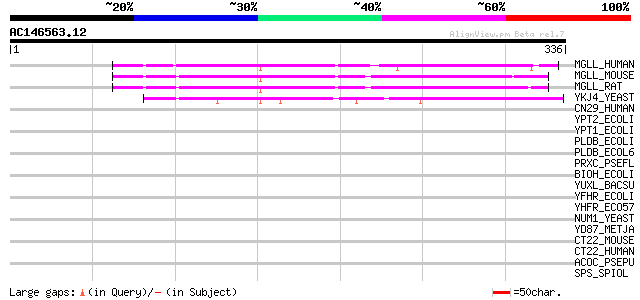
(336 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MGLL_HUMAN (Q99685) Monoglyceride lipase (EC 3.1.1.23) (HU-K5) (... 100 4e-21
MGLL_MOUSE (O35678) Monoglyceride lipase (EC 3.1.1.23) 100 9e-21
MGLL_RAT (Q8R431) Monoglyceride lipase (EC 3.1.1.23) 96 2e-19
YKJ4_YEAST (P28321) Hypothetical 35.5 kDa protein in CWP1-MBR1 i... 74 7e-13
CN29_HUMAN (Q7Z5M8) Protein C14orf29 43 0.001
YPT2_ECOLI (Q99390) Hypothetical 31.7 kDa protein in traX-finO i... 42 0.002
YPT1_ECOLI (P29368) Hypothetical 31.7 kDa protein in traX-finO i... 42 0.002
PLDB_ECOLI (P07000) Lysophospholipase L2 (EC 3.1.1.5) (Lecithina... 41 0.005
PLDB_ECOL6 (P59588) Lysophospholipase L2 (EC 3.1.1.5) (Lecithina... 41 0.005
PRXC_PSEFL (O31158) Non-heme chloroperoxidase (EC 1.11.1.10) (Ch... 37 0.091
BIOH_ECOLI (P13001) BioH protein 35 0.35
YUXL_BACSU (P39839) Probable peptidase yuxL (EC 3.4.21.-) 34 0.45
YFHR_ECOLI (P77538) Hypothetical protein yfhR 34 0.45
YHFR_ECO57 (Q8XA81) Hypothetical protein yfhR 34 0.59
NUM1_YEAST (Q00402) Nuclear migration protein NUM1 33 1.3
YD87_METJA (Q58782) Hypothetical UPF0272 protein MJ1387 32 1.7
CT22_MOUSE (Q99LR1) Protein C20orf22 homolog 32 1.7
CT22_HUMAN (Q8N2K0) Protein C20orf22 32 1.7
ACOC_PSEPU (Q59695) Dihydrolipoyllysine-residue acetyltransferas... 32 2.3
SPS_SPIOL (P31928) Sucrose-phosphate synthase (EC 2.4.1.14) (UDP... 31 5.0
>MGLL_HUMAN (Q99685) Monoglyceride lipase (EC 3.1.1.23) (HU-K5)
(Lysophospholipase homolog) (Lysophospholipase-like)
Length = 303
Score = 100 bits (250), Expect = 4e-21
Identities = 82/285 (28%), Positives = 135/285 (46%), Gaps = 26/285 (9%)
Query: 63 VNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGF 122
VN+ G +F + W P +P K +++ HG + Y E +AR L VFA D+ G
Sbjct: 23 VNADGQYLFCRYWKPTGTP-KALIFVSHGAGEHSGRY-EELARMLMGLDLLVFAHDHVGH 80
Query: 123 GLSDGLHGYIPSFENLVNDVIEHFSKIK-----------GESMGGAIALNIHFKQPTAWD 171
G S+G + F V DV++H ++ G SMGGAIA+ ++P +
Sbjct: 81 GQSEGERMVVSDFHVFVRDVLQHVDSMQKDYPGLPVFLLGHSMGGAIAILTAAERPGHFA 140
Query: 172 GAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPY 231
G LI+PL + + V + + VLP L P V K E+ Y
Sbjct: 141 GMVLISPLVLANPESATTFKVLAAKV-LNLVLPNLSLGPIDSSVLSR-----NKTEVDIY 194
Query: 232 NV--LFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA 289
N L + ++ ++LL A +E+ L ++++P L++ G AD + D + L + A
Sbjct: 195 NSDPLICRAGLKVCFGIQLLNAVSRVERALPKLTVPFLLLQGSADRLCDSKGAYLLMELA 254
Query: 290 KVKDKKLCLYKDAFHTLLEGEPDET--IFHVLDDIISWLDDHSST 332
K +DK L +Y+ A+H L + P+ T +FH +I W+ ++T
Sbjct: 255 KSQDKTLKIYEGAYHVLHKELPEVTNSVFH---EINMWVSQRTAT 296
>MGLL_MOUSE (O35678) Monoglyceride lipase (EC 3.1.1.23)
Length = 303
Score = 99.8 bits (247), Expect = 9e-21
Identities = 77/275 (28%), Positives = 133/275 (48%), Gaps = 18/275 (6%)
Query: 63 VNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGF 122
VN+ G +F + W P +P K +++ HG + C Y E +A L VFA D+ G
Sbjct: 23 VNADGQYLFCRYWKPSGTP-KALIFVSHGAGEHCGRYDE-LAHMLKGLDMLVFAHDHVGH 80
Query: 123 GLSDGLHGYIPSFENLVNDVIEHFSKIK-----------GESMGGAIALNIHFKQPTAWD 171
G S+G + F+ V DV++H I+ G SMGGAI++ + ++PT +
Sbjct: 81 GQSEGERMVVSDFQVFVRDVLQHVDTIQKDYPDVPIFLLGHSMGGAISILVAAERPTYFS 140
Query: 172 GAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPY 231
G LI+PL + V + + VLP L V + R+ + +L
Sbjct: 141 GMVLISPLVLANPESASTLKVLAAKL-LNFVLPNMTLGRIDSSV---LSRNKSEVDLYNS 196
Query: 232 NVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKV 291
+ L + ++ ++LL A +E+ + ++LP L++ G AD + D + L + ++
Sbjct: 197 DPLVCRAGLKVCFGIQLLNAVARVERAMPRLTLPFLLLQGSADRLCDSKGAYLLMESSRS 256
Query: 292 KDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWL 326
+DK L +Y+ A+H +L E E VL ++ SW+
Sbjct: 257 QDKTLKMYEGAYH-VLHRELPEVTNSVLHEVNSWV 290
>MGLL_RAT (Q8R431) Monoglyceride lipase (EC 3.1.1.23)
Length = 303
Score = 95.5 bits (236), Expect = 2e-19
Identities = 75/275 (27%), Positives = 133/275 (48%), Gaps = 18/275 (6%)
Query: 63 VNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGF 122
VN+ G +F + W P +P K +++ HG + C Y E +A+ L VFA D+ G
Sbjct: 23 VNADGQYLFCRYWKPSGTP-KALIFVSHGAGEHCGRYDE-LAQMLKRLDMLVFAHDHVGH 80
Query: 123 GLSDGLHGYIPSFENLVNDVIEHFSKIK-----------GESMGGAIALNIHFKQPTAWD 171
G S+G + F+ V D+++H + ++ G SMGGAI++ ++PT +
Sbjct: 81 GQSEGERMVVSDFQVFVRDLLQHVNTVQKDYPEVPVFLLGHSMGGAISILAAAERPTHFS 140
Query: 172 GAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPY 231
G LI+PL + V + + VLP L V + R+ + +L
Sbjct: 141 GMILISPLILANPESASTLKVLAAKL-LNFVLPNISLGRIDSSV---LSRNKSEVDLYNS 196
Query: 232 NVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKV 291
+ L ++ ++LL A +E+ + ++LP L++ G AD + D + L + +
Sbjct: 197 DPLICHAGVKVCFGIQLLNAVSRVERAMPRLTLPFLLLQGSADRLCDSKGAYLLMESSPS 256
Query: 292 KDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWL 326
+DK L +Y+ A+H L + P+ T VL +I +W+
Sbjct: 257 QDKTLKMYEGAYHVLHKELPEVT-NSVLHEINTWV 290
>YKJ4_YEAST (P28321) Hypothetical 35.5 kDa protein in CWP1-MBR1
intergenic region
Length = 313
Score = 73.6 bits (179), Expect = 7e-13
Identities = 72/280 (25%), Positives = 120/280 (42%), Gaps = 32/280 (11%)
Query: 82 MKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGL-----SDGLHGYIPSFE 136
++G V HG+ + F + L+ +G+ F D G G+ S G+ F
Sbjct: 39 VRGRVLLIHGFGEYTKIQFR-LMDHLSLNGYESFTFDQRGAGVTSPGRSKGVTDEYHVFN 97
Query: 137 NLVNDVIEHFSKIK---------GESMGGAIALNI-----HFKQPTAWDGAALIAPLCKF 182
+L + V ++ S+ K G SMGG I LN H + + + G+ + L
Sbjct: 98 DLEHFVEKNLSECKAKGIPLFMWGHSMGGGICLNYACQGKHKNEISGYIGSGPLIILHPH 157
Query: 183 AEDMIPHWLVKQILIGVAKVLPKTKL---VPQKEEVKENIYRDARKRELAPYNVLFYKDK 239
P ++ +L AK LP+ ++ + K + YR + P +V Y
Sbjct: 158 TMYNKPTQIIAPLL---AKFLPRVRIDTGLDLKGITSDKAYRAFLGSD--PMSVPLYGSF 212
Query: 240 PRLGTALE----LLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKK 295
++ ++ L K Q+ P+++MHG+ D I DP S+ Q DK+
Sbjct: 213 RQIHDFMQRGAKLYKNENNYIQKNFAKDKPVVIMHGQDDTINDPKGSEKFIQDCPSADKE 272
Query: 296 LCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
L LY A H++ E DE V +D+ WLD H++T+ K
Sbjct: 273 LKLYPGARHSIFSLETDEVFNTVFNDMKQWLDKHTTTEAK 312
>CN29_HUMAN (Q7Z5M8) Protein C14orf29
Length = 362
Score = 42.7 bits (99), Expect = 0.001
Identities = 63/251 (25%), Positives = 105/251 (41%), Gaps = 44/251 (17%)
Query: 85 IVYYCHGYAD-TCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVI 143
I+ Y HG A+ + + + L+ GF V ++DY GFG S G P+ E L D I
Sbjct: 141 IIVYLHGSAEHRAASHRLKLVKVLSDGGFHVLSVDYRGFGDSTGK----PTEEGLTTDAI 196
Query: 144 EHFSKIKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVL 203
+ K S I P+C + H L + AKVL
Sbjct: 197 CVYEWTKARSG---------------------ITPVCLWG-----HSLGTGVATNAAKVL 230
Query: 204 PKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPR-LGTALELLKATQEL---EQRL 259
+ K P V E + + + + Y++ P L T ++ L+ + + ++ +
Sbjct: 231 EE-KGCPVDAIVLEAPFTNMWVASINYPLLKIYRNIPGFLRTLMDALRKDKIIFPNDENV 289
Query: 260 EEVSLPLLVMHGEADIITDPSASKALYQKAK--VKDK---KLCLYKDAF-HTLLEGEPDE 313
+ +S PLL++HGE D K LY+ A+ ++K K+ ++ F H LL P
Sbjct: 290 KFLSSPLLILHGEDDRTVPLEYGKKLYEIARNAYRNKERVKMVIFPPGFQHNLLCKSP-- 347
Query: 314 TIFHVLDDIIS 324
T+ + D +S
Sbjct: 348 TLLITVRDFLS 358
>YPT2_ECOLI (Q99390) Hypothetical 31.7 kDa protein in traX-finO
intergenic region (ORFC)
Length = 286
Score = 42.0 bits (97), Expect = 0.002
Identities = 62/267 (23%), Positives = 94/267 (34%), Gaps = 60/267 (22%)
Query: 76 IPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHG-YIPS 134
+PE + ++ CHG+ A +GF DY GFG SDG G +P+
Sbjct: 18 VPEGNIKHPLIILCHGFCGIRNVLLPCFANAFTEAGFATITFDYRGFGESDGERGRLVPA 77
Query: 135 FENL-VNDVIEHFSK----------IKGESMGG-----AIALNIHFK----QPTAWDG-- 172
+ + VI K + G S+GG A A + K Q DG
Sbjct: 78 MQTEDIISVINWAEKQECIDNQRIGLWGTSLGGGHVFSAAAQDQRVKCIVSQLAFADGDV 137
Query: 173 -----------AALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYR 221
A+ ++ L K AE K++ +GV +VL + E+VK
Sbjct: 138 LVTGEMNESERASFLSTLNKMAEK--KKNTGKEMFVGVTRVLSDNESKVFFEKVKGQY-- 193
Query: 222 DARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEE----VSLPLLVMHGEADIIT 277
P + + L + L+ + E V P+L++ D +
Sbjct: 194 ------------------PEMDIKIPFLTVMETLQYKPAESAAKVQCPVLIVIAGQDSVN 235
Query: 278 DPSASKALYQKAKVKDKKLCLYKDAFH 304
P KALY K+L DA H
Sbjct: 236 PPEQGKALYDAVASGTKELYEEADACH 262
>YPT1_ECOLI (P29368) Hypothetical 31.7 kDa protein in traX-finO
intergenic region
Length = 286
Score = 42.0 bits (97), Expect = 0.002
Identities = 59/267 (22%), Positives = 91/267 (33%), Gaps = 60/267 (22%)
Query: 76 IPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHG-YIPS 134
+PE + ++ CHG+ A +GF DY GFG SDG G +P+
Sbjct: 18 VPEGNIKHPLIILCHGFCGIRNVLLPCFANAFTEAGFATITFDYRGFGESDGERGRLVPA 77
Query: 135 FENL-VNDVIEHFSK----------IKGESMGGAIALNIH---------FKQPTAWDG-- 172
+ + VI K + G S+GG + Q DG
Sbjct: 78 MQTEDIISVINWAEKQECIDNQRIGLWGTSLGGGHVFSARAQDQRVKCIVSQLAFADGDV 137
Query: 173 -----------AALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYR 221
A+ ++ L K AE K++ +GV +VL + E+VK
Sbjct: 138 LVTGEMNESERASFLSTLNKMAEK--KKNTGKEMFVGVTRVLSDNESKVFFEKVKGQY-- 193
Query: 222 DARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEE----VSLPLLVMHGEADIIT 277
P + + L + L+ + E V P+L++ D +
Sbjct: 194 ------------------PEMDIKIPFLTVMETLQYKPAESAAKVQCPVLIVIAGQDSVN 235
Query: 278 DPSASKALYQKAKVKDKKLCLYKDAFH 304
P KALY K+L DA H
Sbjct: 236 PPEQGKALYDAVASGTKELYEEADACH 262
>PLDB_ECOLI (P07000) Lysophospholipase L2 (EC 3.1.1.5) (Lecithinase
B)
Length = 340
Score = 40.8 bits (94), Expect = 0.005
Identities = 67/285 (23%), Positives = 115/285 (39%), Gaps = 47/285 (16%)
Query: 85 IVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFG-----LSDGLHGYIPSFENLV 139
+V C G ++ Y E +A L GF V +D+ G G L+D G++ F + V
Sbjct: 56 VVVICPGRIESYVKYAE-LAYDLFHLGFDVLIIDHRGQGRSGRLLADPHLGHVNRFNDYV 114
Query: 140 NDVIEHFSK-----------IKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIP 188
+D+ + + I SMGGAI+ + P D AL AP+ M P
Sbjct: 115 DDLAAFWQQEVQPGPWRKRYILAHSMGGAISTLFLQRHPGVCDAIALTAPMFGIVIRM-P 173
Query: 189 HWLVKQIL----------IGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD 238
++ +QIL G A + + +P N+ +R+R N+ FY D
Sbjct: 174 SFMARQILNWAEAHPRFRDGYAIGTGRWRALP----FAINVLTHSRQRYRR--NLRFYAD 227
Query: 239 KP--RLG-----TALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA-- 289
P R+G E + A +++ + + P L++ E + + D +
Sbjct: 228 DPTIRVGGPTYHWVRESILAGEQVLAGAGDDATPTLLLQAEEERVVDNRMHDRFCELRTA 287
Query: 290 ---KVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSS 331
V+ + + K A+H +L E D L I+ + + H+S
Sbjct: 288 AGHPVEGGRPLVIKGAYHEIL-FEKDAMRSVALHAIVDFFNRHNS 331
>PLDB_ECOL6 (P59588) Lysophospholipase L2 (EC 3.1.1.5) (Lecithinase
B)
Length = 340
Score = 40.8 bits (94), Expect = 0.005
Identities = 67/285 (23%), Positives = 115/285 (39%), Gaps = 47/285 (16%)
Query: 85 IVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFG-----LSDGLHGYIPSFENLV 139
+V C G ++ Y E +A L GF V +D+ G G L+D G++ F + V
Sbjct: 56 VVVICPGRIESYVKYAE-LAYDLFHLGFDVLIIDHRGQGRSGRLLADPHLGHVNRFNDYV 114
Query: 140 NDVIEHFSK-----------IKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIP 188
+D+ + + I SMGGAI+ + P D AL AP+ M P
Sbjct: 115 DDLAAFWQQEVQPGPWRKRYILAHSMGGAISTLFLQRHPGVCDAIALTAPMFGIVIRM-P 173
Query: 189 HWLVKQIL----------IGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD 238
++ +QIL G A + + +P N+ +R+R N+ FY D
Sbjct: 174 SFMARQILNWAEAHPRFRDGYAIGTGRWRALP----FAINVLTHSRQRYRR--NLRFYAD 227
Query: 239 KP--RLG-----TALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA-- 289
P R+G E + A +++ + + P L++ E + + D +
Sbjct: 228 DPTIRVGGPTYHWVRESILAGEQVLAGAGDDATPTLLLQAEEERVVDNRMHDRFCELRTA 287
Query: 290 ---KVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSS 331
V+ + + K A+H +L E D L I+ + + H+S
Sbjct: 288 AGHPVEGGRPLVIKGAYHEIL-FEKDAMRSVALHAIVDFFNRHNS 331
>PRXC_PSEFL (O31158) Non-heme chloroperoxidase (EC 1.11.1.10)
(Chloride peroxidase) (CPO-F) (Chloroperoxidase F)
Length = 273
Score = 36.6 bits (83), Expect = 0.091
Identities = 22/71 (30%), Positives = 38/71 (52%), Gaps = 3/71 (4%)
Query: 245 ALELLKATQELE--QRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDA 302
A + +KA E + + L+++ +P LV+HG+AD + P + + A VK L +Y A
Sbjct: 193 AYDCIKAFSETDFTEDLKKIDVPTLVVHGDADQVV-PIEASGIASAALVKGSTLKIYSGA 251
Query: 303 FHTLLEGEPDE 313
H L + D+
Sbjct: 252 PHGLTDTHKDQ 262
>BIOH_ECOLI (P13001) BioH protein
Length = 256
Score = 34.7 bits (78), Expect = 0.35
Identities = 47/176 (26%), Positives = 71/176 (39%), Gaps = 15/176 (8%)
Query: 109 SSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK---GESMGGAIALNIHFK 165
SS F + +D PGFG S G S ++ V++ G S+GG +A I
Sbjct: 37 SSHFTLHLVDLPGFGRSRGFGAL--SLADMAEAVLQQAPDKAIWLGWSLGGLVASQIALT 94
Query: 166 QPTAWDGAALIAPL-CKFAEDMIPHWLVKQILIGVAKVLPK------TKLVPQKEEVKEN 218
P +A C A D P + +L G + L + + + E
Sbjct: 95 HPERVQALVTVASSPCFSARDEWP-GIKPDVLAGFQQQLSDDFQRTVERFLALQTMGTET 153
Query: 219 IYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEAD 274
+DAR + L + L LE+LK T +L Q L+ VS+P L ++G D
Sbjct: 154 ARQDARALKKTVL-ALPMPEVDVLNGGLEILK-TVDLRQPLQNVSMPFLRLYGYLD 207
>YUXL_BACSU (P39839) Probable peptidase yuxL (EC 3.4.21.-)
Length = 657
Score = 34.3 bits (77), Expect = 0.45
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query: 262 VSLPLLVMHGEADIITDPSASKALY--QKAKVKDKKLCLYKDAFHTLLE-GEPDETIFHV 318
V PLL++HGE D ++ L+ K K+ KL + +A H L G P + I
Sbjct: 587 VETPLLILHGERDDRCPIEQAEQLFIALKKMGKETKLVRFPNASHNLSRTGHPRQRIKR- 645
Query: 319 LDDIISWLDDH 329
L+ I SW D H
Sbjct: 646 LNYISSWFDQH 656
>YFHR_ECOLI (P77538) Hypothetical protein yfhR
Length = 284
Score = 34.3 bits (77), Expect = 0.45
Identities = 26/87 (29%), Positives = 42/87 (47%), Gaps = 12/87 (13%)
Query: 86 VYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGL---HGYIPSFENLVNDV 142
+ + HG A + ++ V+ L F VF DY GFG S G G + ++ +N V
Sbjct: 81 IIHAHGNAGNMSAHWPLVSW-LPERNFNVFMFDYRGFGKSKGTPSQAGLLDDTQSAIN-V 138
Query: 143 IEHFSKIK-------GESMGGAIALNI 162
+ H S + G+S+GGA L++
Sbjct: 139 VRHRSDVNPQRLVLFGQSIGGANILDV 165
>YHFR_ECO57 (Q8XA81) Hypothetical protein yfhR
Length = 284
Score = 33.9 bits (76), Expect = 0.59
Identities = 57/221 (25%), Positives = 86/221 (38%), Gaps = 47/221 (21%)
Query: 86 VYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEH 145
+ + HG A + ++ V+ L F VF DY GFG S G PS L++D
Sbjct: 81 IIHAHGNAGNMSAHWPLVSW-LPERNFNVFMFDYRGFGKSKGT----PSQAGLLDDTQSA 135
Query: 146 FSKIKGESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVK-QILIGVAKVLP 204
+ ++ S D+ P LV IG A +L
Sbjct: 136 INVVRHRS-------------------------------DVNPQRLVLFGQSIGGANILA 164
Query: 205 KTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVS- 263
++ Q + +E I A Y + + P G LL + E + VS
Sbjct: 165 ---VIGQGD--REGIRAVILDSTFASYATIANQMIPGSGY---LLDESYSGENYIASVSP 216
Query: 264 LPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYKDAFH 304
+PLL++HG+AD + S+ LY AK + K+L L D H
Sbjct: 217 IPLLLIHGKADHVIPWQHSEKLYSLAK-EPKRLILIPDGEH 256
>NUM1_YEAST (Q00402) Nuclear migration protein NUM1
Length = 2748
Score = 32.7 bits (73), Expect = 1.3
Identities = 36/127 (28%), Positives = 64/127 (50%), Gaps = 9/127 (7%)
Query: 215 VKENIYRDARKRELAPYNVLFYKDKP-RLGTALELLKATQELEQRLEEVSLPLLVMHGEA 273
+ ++ Y D K + P +V F K+K +LG + +A ELE++LE+ SL LV H +A
Sbjct: 1200 LSDSAYEDLVKCKENP-DVEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLAYLVEHAKA 1258
Query: 274 ---DIITDPSASKALYQKAKVKDKKLCLYKDA--FHTLLEGEPDETIFHVLDD-IISWLD 327
+++D SA + L + + D + K A HT++ E + L+ + +L
Sbjct: 1259 TDHHLLSD-SAYEDLVKCKENPDMEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLEYLV 1317
Query: 328 DHSSTKN 334
+H+ N
Sbjct: 1318 EHAKATN 1324
Score = 31.6 bits (70), Expect = 2.9
Identities = 35/127 (27%), Positives = 64/127 (49%), Gaps = 9/127 (7%)
Query: 215 VKENIYRDARKRELAPYNVLFYKDKP-RLGTALELLKATQELEQRLEEVSLPLLVMHGEA 273
+ ++ Y D K + P ++ F K+K +LG + +A ELE++LE+ SL LV H +A
Sbjct: 944 LSDSAYEDLVKCKENP-DMEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLEYLVEHAKA 1002
Query: 274 ---DIITDPSASKALYQKAKVKDKKLCLYKDA--FHTLLEGEPDETIFHVLDD-IISWLD 327
+++D SA + L + + D + K A HT++ E + L+ + +L
Sbjct: 1003 TNHHLLSD-SAYEDLVKCKENPDMEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLEYLV 1061
Query: 328 DHSSTKN 334
+H+ N
Sbjct: 1062 EHAKATN 1068
Score = 30.8 bits (68), Expect = 5.0
Identities = 32/110 (29%), Positives = 56/110 (50%), Gaps = 8/110 (7%)
Query: 232 NVLFYKDKP-RLGTALELLKATQELEQRLEEVSLPLLVMHGEA---DIITDPSASKALYQ 287
+V F K+K +LG + +A ELE++LE+ SL LV H +A +++D SA + L +
Sbjct: 896 DVEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLAYLVEHAKATDHHLLSD-SAYEDLVK 954
Query: 288 KAKVKDKKLCLYKDA--FHTLLEGEPDETIFHVLDD-IISWLDDHSSTKN 334
+ D + K A HT++ E + L+ + +L +H+ N
Sbjct: 955 CKENPDMEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLEYLVEHAKATN 1004
Score = 30.0 bits (66), Expect = 8.6
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query: 208 LVPQKEEVKENIYRDARKRELAPY----NVLFYKDKP-RLGTALELLKATQELEQRLEEV 262
LV + ++ D+ EL +V F K+K +LG + +A ELE++LE+
Sbjct: 1060 LVEHAKATNHHLLSDSAYEELVKCKENPDVEFLKEKSAKLGHTVVSNEAYSELEKKLEQP 1119
Query: 263 SLPLLVMHGEA 273
SL LV H +A
Sbjct: 1120 SLEYLVEHAKA 1130
Score = 30.0 bits (66), Expect = 8.6
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query: 208 LVPQKEEVKENIYRDARKRELAPY----NVLFYKDKP-RLGTALELLKATQELEQRLEEV 262
LV + ++ D+ EL +V F K+K +LG + +A ELE++LE+
Sbjct: 1124 LVEHAKATNHHLLSDSAYEELVKCKENPDVEFLKEKSAKLGHTVVSNEAYSELEKKLEQP 1183
Query: 263 SLPLLVMHGEA 273
SL LV H +A
Sbjct: 1184 SLAYLVEHAKA 1194
Score = 30.0 bits (66), Expect = 8.6
Identities = 21/60 (35%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query: 215 VKENIYRDARKRELAPYNVLFYKDKP-RLGTALELLKATQELEQRLEEVSLPLLVMHGEA 273
+ ++ Y D K + P ++ F K+K +LG + +A ELE++LE+ SL LV H +A
Sbjct: 610 LSDSAYEDLVKCKENP-DMEFLKEKSAKLGHTVVSNEAYSELEKKLEQPSLEYLVEHAKA 668
Score = 30.0 bits (66), Expect = 8.6
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query: 215 VKENIYRDARKRELAPYNVLFYKDKP-RLGTALELLKATQELEQRLEEVSLPLLVMHGE 272
+ ++ Y D K + P ++ F K+K +LG + K ELE++LE+ SL LV H E
Sbjct: 1328 LSDSAYEDLVKCKENP-DMEFLKEKSAKLGHTVVSNKEYSELEKKLEQPSLEYLVKHAE 1385
>YD87_METJA (Q58782) Hypothetical UPF0272 protein MJ1387
Length = 379
Score = 32.3 bits (72), Expect = 1.7
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 13/100 (13%)
Query: 205 KTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD--KPRLGTALELLKATQELEQRLEEV 262
K ++P+ N Y+D + NV+ D + TALE+LK E E ++ V
Sbjct: 54 KVNIIPKCINCNANTYKDIK-------NVIKSSDIQEDIKITALEILKILAEAESKVHNV 106
Query: 263 SLPLLVMH--GEADIITDPSASKALYQKAKVKDKKLCLYK 300
+ + H G D I D + + K +K+ CLYK
Sbjct: 107 DVENVHFHEVGNYDTIADIVGAAYIINKLNLKNN--CLYK 144
>CT22_MOUSE (Q99LR1) Protein C20orf22 homolog
Length = 398
Score = 32.3 bits (72), Expect = 1.7
Identities = 55/221 (24%), Positives = 84/221 (37%), Gaps = 58/221 (26%)
Query: 85 IVYYCHGYADTCTFYFEGVARKLASS-GFGVFALDYPGFGLSDGLHGYIPSFENLVNDVI 143
I+ Y HG A T K+ SS G+ V DY G+G S G PS + D +
Sbjct: 170 IILYLHGNAGTRGGDHRVELYKVLSSLGYHVVTFDYRGWGDSVG----TPSERGMTYDAL 225
Query: 144 EHFSKIK-----------GESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLV 192
F IK G S+G +A N L+ LC+ + P L+
Sbjct: 226 HVFDWIKARSGDNPVYIWGHSLGTGVATN-------------LVRRLCE--RETPPDALI 270
Query: 193 KQILIGVAKVLPKTKLVPQKEEVKEN----IYRDARKRELAPYNVLFYKDKPRLGTALEL 248
+ P T + +EE K + IYR P F+ D P + ++
Sbjct: 271 LE--------SPFTNI---REEAKSHPFSVIYR------YFPGFDWFFLD-PITSSGIKF 312
Query: 249 LKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA 289
++ ++ +S PLL++H E D + + LY A
Sbjct: 313 AN-----DENMKHISCPLLILHAEDDPVVPFHLGRKLYNIA 348
>CT22_HUMAN (Q8N2K0) Protein C20orf22
Length = 398
Score = 32.3 bits (72), Expect = 1.7
Identities = 55/221 (24%), Positives = 84/221 (37%), Gaps = 58/221 (26%)
Query: 85 IVYYCHGYADTCTFYFEGVARKLASS-GFGVFALDYPGFGLSDGLHGYIPSFENLVNDVI 143
I+ Y HG A T K+ SS G+ V DY G+G S G PS + D +
Sbjct: 170 IILYLHGNAGTRGGDHRVELYKVLSSLGYHVVTFDYRGWGDSVG----TPSERGMTYDAL 225
Query: 144 EHFSKIK-----------GESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLV 192
F IK G S+G +A N L+ LC+ + P L+
Sbjct: 226 HVFDWIKARSGDNPVYIWGHSLGTGVATN-------------LVRRLCE--RETPPDALI 270
Query: 193 KQILIGVAKVLPKTKLVPQKEEVKEN----IYRDARKRELAPYNVLFYKDKPRLGTALEL 248
+ P T + +EE K + IYR P F+ D P + ++
Sbjct: 271 LE--------SPFTNI---REEAKSHPFSVIYR------YFPGFDWFFLD-PITSSGIKF 312
Query: 249 LKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKA 289
++ ++ +S PLL++H E D + + LY A
Sbjct: 313 AN-----DENVKHISCPLLILHAEDDPVVPFQLGRKLYSIA 348
>ACOC_PSEPU (Q59695) Dihydrolipoyllysine-residue acetyltransferase
component of acetoin cleaving system (EC 2.3.1.12)
(Acetoin dehydrogenase E2 component) (Dihydrolipoamide
acetyltransferase component of acetoin cleaving system)
Length = 370
Score = 32.0 bits (71), Expect = 2.3
Identities = 24/70 (34%), Positives = 32/70 (45%), Gaps = 6/70 (8%)
Query: 114 VFALDYPGFGLSDGL--HGYIPSFENLVNDVIEHFSKIK----GESMGGAIALNIHFKQP 167
V ALD PG G S G + V +++H K G SMGGA++LN+ P
Sbjct: 162 VIALDLPGHGESAKALQRGDLDELSETVLALLDHLDIAKAHLAGHSMGGAVSLNVAGLAP 221
Query: 168 TAWDGAALIA 177
+LIA
Sbjct: 222 QRVASLSLIA 231
>SPS_SPIOL (P31928) Sucrose-phosphate synthase (EC 2.4.1.14)
(UDP-glucose-fructose-phosphate glucosyltransferase)
Length = 1056
Score = 30.8 bits (68), Expect = 5.0
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Query: 131 YIPSFENLVNDVIEHFSKIKGESMGGAIAL---NIHFKQPTAWDGAALIA 177
YIP F + I+ SK+ GE +GG + + ++H A D AAL++
Sbjct: 284 YIPEFVDGALSHIKQMSKVLGEQIGGGLPVWPASVHGHYADAGDSAALLS 333
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,918,182
Number of Sequences: 164201
Number of extensions: 1774993
Number of successful extensions: 4500
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 4471
Number of HSP's gapped (non-prelim): 48
length of query: 336
length of database: 59,974,054
effective HSP length: 111
effective length of query: 225
effective length of database: 41,747,743
effective search space: 9393242175
effective search space used: 9393242175
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146563.12


