

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.10 + phase: 0
(231 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
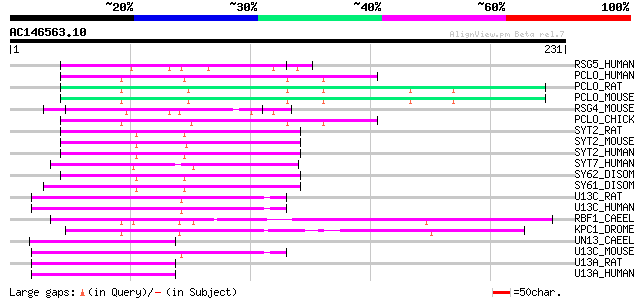
Score E
Sequences producing significant alignments: (bits) Value
RSG5_HUMAN (O43374) Ras GTPase-activating protein 4 (RasGAP-acti... 64 2e-10
PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin) 58 2e-08
PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytom... 57 5e-08
PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix prot... 55 1e-07
RSG4_MOUSE (Q9Z268) RasGAP-activating-like protein 1 54 2e-07
PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment) 54 2e-07
SYT2_RAT (P29101) Synaptotagmin-2 (Synaptotagmin II) (SytII) 53 7e-07
SYT2_MOUSE (P46097) Synaptotagmin-2 (Synaptotagmin II) (SytII) 53 7e-07
SYT2_HUMAN (Q8N9I0) Synaptotagmin-2 (Synaptotagmin II) (SytII) 53 7e-07
SYT7_HUMAN (O43581) Synaptotagmin-7 (Synaptotagmin VII) (SytVII) 52 9e-07
SY62_DISOM (P24506) Synaptotagmin B (Synaptic vesicle protein O-... 52 9e-07
SY61_DISOM (P24505) Synaptotagmin A (Synaptic vesicle protein O-... 52 9e-07
U13C_RAT (Q62770) Unc-13 homolog C (Munc13-3) 51 2e-06
U13C_HUMAN (Q8NB66) Unc-13 homolog C (Munc13-3) (Fragment) 51 2e-06
RBF1_CAEEL (P41885) Rabphilin 1 51 2e-06
KPC1_DROME (P05130) Protein kinase C, brain isozyme (EC 2.7.1.-)... 51 2e-06
UN13_CAEEL (P27715) Phorbol ester/diacylglycerol-binding protein... 51 3e-06
U13C_MOUSE (Q8K0T7) Unc-13 homolog C (Munc13-3) (Fragment) 51 3e-06
U13A_RAT (Q62768) Unc-13 homolog A (Munc13-1) 51 3e-06
U13A_HUMAN (Q9UPW8) Unc-13 homolog A (Munc13-1) (Fragment) 51 3e-06
>RSG5_HUMAN (O43374) Ras GTPase-activating protein 4
(RasGAP-activating-like protein 2) (Calcium-promoted Ras
inactivator)
Length = 803
Score = 64.3 bits (155), Expect = 2e-10
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPLMLYV 80
++E RDL D GTSDP+VRV Y + T ++ K+ P+WN+T EF +G+ L V
Sbjct: 139 VLEARDLAPKDRNGTSDPFVRVRYKGRTRETSIVKKSCYPRWNETFEFELQEGAMEALCV 198
Query: 81 K--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
+ D + + +G+ V++ QRL Q + W LQ
Sbjct: 199 EAWDWDLVSRNDFLGKVVIDVQRLRVVQQEEGWFRLQ 235
Score = 47.4 bits (111), Expect = 3e-05
Identities = 34/114 (29%), Positives = 54/114 (46%), Gaps = 9/114 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFK-KRTKVIYKTLTPQWNQ--TLEFPDDGSPLML 78
++EG++L A D+ G+SDPY V N RT ++KTL P W + + P +
Sbjct: 11 IVEGKNLPAKDITGSSDPYCIVKVDNEPIIRTATVWKTLCPFWGEEYQVHLPPTFHAVAF 70
Query: 79 YVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGVK-----RGEIHIQI 126
YV D +AL IG+ + + + W L V +GEIH+++
Sbjct: 71 YVMDEDALSRDDVIGKVCLTRDTIASHPKGFSGWAHLTEVDPDEEVQGEIHLRL 124
>PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin)
Length = 5183
Score = 57.8 bits (138), Expect = 2e-08
Identities = 40/150 (26%), Positives = 70/150 (46%), Gaps = 18/150 (12%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHY-----GNFKKRTKVIYKTLTPQWNQTLEFPD----- 71
+++ R+LV D G SDP+V+V+ +K+RTK + K+L P+WNQT+ +
Sbjct: 4584 ILQARNLVPRDNNGYSDPFVKVYLLPGRGAEYKRRTKHVQKSLNPEWNQTVIYKSISMEQ 4643
Query: 72 -DGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL----QGVKRGEIHIQI 126
L + V D++ +GE +++ +W PL + + G+ H
Sbjct: 4644 LKKKTLEVTVWDYDRFSSNDFLGEVLIDLSSTSHLDNTPRWYPLKEQTESIDHGKSHSSQ 4703
Query: 127 TRK---VPEMQKRQSMDSEPSLSKLHQIPT 153
+ + P + K +S P SK Q+PT
Sbjct: 4704 SSQQSPKPSVIKSRSHGIFPDPSKDMQVPT 4733
>PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytomatrix
protein)
Length = 5085
Score = 56.6 bits (135), Expect = 5e-08
Identities = 57/236 (24%), Positives = 93/236 (39%), Gaps = 34/236 (14%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHY--------------GNFKKRTKVIYKTLTPQWNQTL 67
+++ R+LV D G SDP+V+V+ +K+RTK + K+L P+WNQT+
Sbjct: 4658 ILQARNLVPRDNNGYSDPFVKVYLLPGRGQVMVVQNASAEYKRRTKYVQKSLNPEWNQTV 4717
Query: 68 EFPDDG------SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL----QGV 117
+ L + V D++ +GE +++ +W PL + +
Sbjct: 4718 IYKSISMEQLMKKTLEVTVWDYDRFSSNDFLGEVLIDLSSTSHLDNTPRWYPLKEQTESI 4777
Query: 118 KRGEIHIQITRK---VPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQ-IEDGNLEG 173
G+ H + P + K +S P SK Q+PT K S+ +G+L
Sbjct: 4778 DHGKSHSSQNSQQSPKPSVIKSRSHGIFPDPSKDMQVPTIEKSHSSPGSSKSSSEGHLRS 4837
Query: 174 LSTTLSELET------LEDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLS 223
+ S+ +T LED A Q L + K SS S S
Sbjct: 4838 HGPSRSQSKTSVAQTHLEDAGVAIAAAEAAVQQLRIQPTKPTNHRPAESSVSTGSS 4893
>PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix protein)
(Aczonin) (Brain-derived HLMN protein)
Length = 5038
Score = 55.5 bits (132), Expect = 1e-07
Identities = 56/236 (23%), Positives = 94/236 (39%), Gaps = 34/236 (14%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHY--------------GNFKKRTKVIYKTLTPQWNQTL 67
+++ R+LV D G SDP+V+V+ +K+RTK + K+L P+WNQT+
Sbjct: 4611 ILQARNLVPRDNNGYSDPFVKVYLLPGRGQVMVVQNASVEYKRRTKYVQKSLNPEWNQTV 4670
Query: 68 EFPDDG------SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL----QGV 117
+ L + V D++ +GE +++ +W PL + +
Sbjct: 4671 IYKSISMEQLMKKTLEVTVWDYDRFSSNDFLGEVLIDLSSTSHLDNTPRWYPLKEQTESI 4730
Query: 118 KRGEIHIQITRK---VPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQ-IEDGNLEG 173
+ G+ H + P + K +S P SK Q+PT K S+ +G+L
Sbjct: 4731 EHGKSHSSQNSQQSPKPSVIKSRSHGIFPDPSKDMQVPTIEKSHSSPGSSKSSSEGHLRS 4790
Query: 174 LSTTLSELET------LEDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLS 223
+ S+ +T LED A Q L + K +S S S
Sbjct: 4791 HGPSRSQSKTSVAQTHLEDAGAAIAAAEAAVQQLRIQPTKPTNHRPAETSVSTGSS 4846
>RSG4_MOUSE (Q9Z268) RasGAP-activating-like protein 1
Length = 799
Score = 54.3 bits (129), Expect = 2e-07
Identities = 33/90 (36%), Positives = 47/90 (51%), Gaps = 10/90 (11%)
Query: 24 EGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF---PDDGSPLMLYV 80
+ RDL D+ GTSDP+ RV +GN T I KT P W++ LE P SPL + +
Sbjct: 141 QARDLAPRDISGTSDPFARVFWGNHSLETSTIKKTRFPHWDEVLELREAPGTTSPLRVEL 200
Query: 81 KDHNALLPTSSIGECVVEY-----QRLPPN 105
D + + +G +VE+ Q+ PPN
Sbjct: 201 WDWDMVGKNDFLG--MVEFTPQTLQQKPPN 228
Score = 48.9 bits (115), Expect = 1e-05
Identities = 33/107 (30%), Positives = 54/107 (49%), Gaps = 4/107 (3%)
Query: 15 NGWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-FKKRTKVIYKTLTPQWNQ--TLEFPD 71
+G + + ++EGR L A D+ G+SDPY V + RT I+++L+P W + T+ P
Sbjct: 4 SGSLSIRVVEGRALPAKDVSGSSDPYCLVKVDDQVVARTATIWRSLSPFWGEEYTVHLPL 63
Query: 72 DGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGV 117
D L YV D + + IG+ + + + + D WI L V
Sbjct: 64 DFHHLAFYVLDEDTVGHDDIIGKISLSKEAITADPRGIDSWINLSRV 110
>PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment)
Length = 5120
Score = 54.3 bits (129), Expect = 2e-07
Identities = 39/159 (24%), Positives = 71/159 (44%), Gaps = 27/159 (16%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHY--------------GNFKKRTKVIYKTLTPQWNQTL 67
+++ R+L D G SDP+V+V+ +K+RTK + K+L P+WNQT+
Sbjct: 4632 ILQARNLAPRDNNGYSDPFVKVYLLPGRGQVMVVQNASAEYKRRTKYVQKSLNPEWNQTV 4691
Query: 68 EFPDDGS------PLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPL----QGV 117
+ + + L + V D++ +GE +++ + +W PL + +
Sbjct: 4692 IYKNISTEQLKKKTLEVTVWDYDRFSSNDFLGEVLIDLSSVSQLDNTPRWYPLKEQSENI 4751
Query: 118 KRGEIHIQITRK---VPEMQKRQSMDSEPSLSKLHQIPT 153
G+ H + P + K +S P SK Q+PT
Sbjct: 4752 DHGKSHSGQNSQQSPKPSVIKSRSHGIFPDPSKDMQVPT 4790
>SYT2_RAT (P29101) Synaptotagmin-2 (Synaptotagmin II) (SytII)
Length = 422
Score = 52.8 bits (125), Expect = 7e-07
Identities = 36/108 (33%), Positives = 54/108 (49%), Gaps = 8/108 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKR---TKVIYKTLTPQWNQTLEFPD-----DG 73
+++ +L A D+ GTSDPYV+V KK+ TKV KTL P +N+T F G
Sbjct: 163 VLQAAELPALDMGGTSDPYVKVFLLPDKKKKYETKVHRKTLNPAFNETFTFKVPYQELGG 222
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
L++ + D + IGE V + Q ++W LQG ++ E
Sbjct: 223 KTLVMAIYDFDRFSKHDIIGEVKVPMNTVDLGQPIEEWRDLQGGEKEE 270
Score = 41.2 bits (95), Expect = 0.002
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 5/59 (8%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-----FKKRTKVIYKTLTPQWNQTLEF 69
G + + ++E ++L D+ G SDPYV++H KK+T V KTL P +N++ F
Sbjct: 288 GKLTVCILEAKNLKKMDVGGLSDPYVKIHLMQNGKRLKKKKTTVKKKTLNPYFNESFSF 346
>SYT2_MOUSE (P46097) Synaptotagmin-2 (Synaptotagmin II) (SytII)
Length = 422
Score = 52.8 bits (125), Expect = 7e-07
Identities = 36/108 (33%), Positives = 54/108 (49%), Gaps = 8/108 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKR---TKVIYKTLTPQWNQTLEFPDD-----G 73
+++ +L A D+ GTSDPYV+V KK+ TKV KTL P +N+T F G
Sbjct: 163 VLQAAELPALDMGGTSDPYVKVFLLPDKKKKYETKVHRKTLNPAFNETFTFKVPYQELAG 222
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
L++ + D + IGE V + Q ++W LQG ++ E
Sbjct: 223 KTLVMAIYDFDRFSKHDIIGEVKVPMNTVDLGQPIEEWRDLQGGEKEE 270
Score = 41.2 bits (95), Expect = 0.002
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 5/59 (8%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-----FKKRTKVIYKTLTPQWNQTLEF 69
G + + ++E ++L D+ G SDPYV++H KK+T V KTL P +N++ F
Sbjct: 288 GKLTVCILEAKNLKKMDVGGLSDPYVKIHLMQNGKRLKKKKTTVKKKTLNPYFNESFSF 346
>SYT2_HUMAN (Q8N9I0) Synaptotagmin-2 (Synaptotagmin II) (SytII)
Length = 419
Score = 52.8 bits (125), Expect = 7e-07
Identities = 36/108 (33%), Positives = 54/108 (49%), Gaps = 8/108 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKR---TKVIYKTLTPQWNQTLEFPD-----DG 73
+++ +L A D+ GTSDPYV+V KK+ TKV KTL P +N+T F G
Sbjct: 160 VLQAAELPALDMGGTSDPYVKVFLLPDKKKKYETKVHRKTLNPAFNETFTFKVPYQELGG 219
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
L++ + D + IGE V + Q ++W LQG ++ E
Sbjct: 220 KTLVMAIYDFDRFSKHDIIGEVKVPMNTVDLGQPIEEWRDLQGGEKEE 267
Score = 38.5 bits (88), Expect = 0.013
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 5/59 (8%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-----FKKRTKVIYKTLTPQWNQTLEF 69
G + + ++E ++L D+ G SDPY ++H KK+T V KTL P +N++ F
Sbjct: 285 GKLTVCILEAKNLKKMDVGGLSDPYGKIHLMQNGKRLKKKKTTVKKKTLNPYFNESFSF 343
>SYT7_HUMAN (O43581) Synaptotagmin-7 (Synaptotagmin VII) (SytVII)
Length = 402
Score = 52.4 bits (124), Expect = 9e-07
Identities = 36/114 (31%), Positives = 55/114 (47%), Gaps = 13/114 (11%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKK---RTKVIYKTLTPQWNQTLEFPDDGS 74
+ L +++ ++L A D GTSDP+V+++ KK +TKV K L P WN+T F +G
Sbjct: 151 LTLKIMKAQELPAKDFSGTSDPFVKIYLLPDKKHKLKTKVKRKNLNPHWNETFLF--EGF 208
Query: 75 P--------LMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRG 120
P L L V D++ IGE + ++ QM W L+ G
Sbjct: 209 PYEKVVQRILYLQVLDYDRFSRNDPIGEVSIPLNKVDLTQMQTFWKDLKPCSDG 262
Score = 38.5 bits (88), Expect = 0.013
Identities = 28/92 (30%), Positives = 50/92 (53%), Gaps = 10/92 (10%)
Query: 22 LIEGRDLVAADLRGTSDPYVRV--HYGN---FKKRTKVIYKTLTPQWNQTLEF--PDD-- 72
+I+ R+L A D+ GTSDPYV+V Y + KK+T + + L P +N++ F P +
Sbjct: 286 IIKARNLKAMDIGGTSDPYVKVWLMYKDKRVEKKKTVTMKRNLNPNFNESFAFDIPTEKL 345
Query: 73 -GSPLMLYVKDHNALLPTSSIGECVVEYQRLP 103
+ +++ V D + L IG+ + ++ P
Sbjct: 346 RETTIIITVMDKDKLSRNDVIGKIYLSWKSGP 377
>SY62_DISOM (P24506) Synaptotagmin B (Synaptic vesicle protein
O-p65-B)
Length = 439
Score = 52.4 bits (124), Expect = 9e-07
Identities = 35/108 (32%), Positives = 54/108 (49%), Gaps = 8/108 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKR---TKVIYKTLTPQWNQTLEFPD-----DG 73
+I+ +L A D+ GTSDPYV+V KK+ TKV KTL P +N++ F G
Sbjct: 180 IIQAAELPALDMGGTSDPYVKVFLLPDKKKKYETKVQKKTLNPTFNESFVFKVPYQELGG 239
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
LM+ V D + IG+ V ++ Q ++W L+ ++ E
Sbjct: 240 KTLMMAVYDFDRFSKHDCIGQVTVLMTKVDLGQQLEEWRDLESAEKEE 287
Score = 39.3 bits (90), Expect = 0.008
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 5/59 (8%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-----FKKRTKVIYKTLTPQWNQTLEF 69
G + + ++E ++L D+ G SDPYV++H KK+T V TL P +N++ F
Sbjct: 305 GKLTVCILEAKNLKKMDVGGLSDPYVKIHLLQNGKRLKKKKTTVKKNTLNPYYNESFSF 363
>SY61_DISOM (P24505) Synaptotagmin A (Synaptic vesicle protein
O-p65-A)
Length = 427
Score = 52.4 bits (124), Expect = 9e-07
Identities = 37/115 (32%), Positives = 57/115 (49%), Gaps = 8/115 (6%)
Query: 15 NGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKR---TKVIYKTLTPQWNQTLEFPD 71
N + + +I+ +L A D+ GTSDPYV+V KK+ TKV KTL P +N++ F
Sbjct: 161 NNQLIVGIIQAAELPALDVGGTSDPYVKVFVLPDKKKKYETKVHRKTLNPVFNESFIFKI 220
Query: 72 -----DGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGE 121
G L++ V D + IGE V + + ++W LQG ++ E
Sbjct: 221 PYSELGGKTLVMAVYDFDRFSKHDVIGEAKVPMNTVDFGHVTEEWRDLQGAEKEE 275
Score = 42.4 bits (98), Expect = 0.001
Identities = 26/94 (27%), Positives = 48/94 (50%), Gaps = 10/94 (10%)
Query: 16 GWIELVLIEGRDLVAADLRGTSDPYVRVHYGN-----FKKRTKVIYKTLTPQWNQTLEFP 70
G + +V++E ++L D+ G SDPYV++H KK+T + TL P +N++ F
Sbjct: 293 GKLTVVILEAKNLKKMDVGGLSDPYVKIHLMQNGKRLKKKKTTIKKNTLNPYYNESFSFE 352
Query: 71 DDGSPL-----MLYVKDHNALLPTSSIGECVVEY 99
+ ++ V D++ + +IG+ V Y
Sbjct: 353 VPFEQIQKVQVVVTVLDYDKIGKNDAIGKVFVGY 386
>U13C_RAT (Q62770) Unc-13 homolog C (Munc13-3)
Length = 2204
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/119 (27%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+ + I + ++ + L A D G+SDPYV V G K+RTK I+ L P W++ F
Sbjct: 1204 GTSKWSAKITITVVSAQGLQAKDKTGSSDPYVTVQVGKNKRRTKTIFGNLNPVWDEKFYF 1263
Query: 70 P-------------DDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
D+ + VK H +G+ +VE + L D W L+
Sbjct: 1264 ECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTLSGEM--DVWYNLE 1320
Score = 29.6 bits (65), Expect = 6.3
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 6/72 (8%)
Query: 49 KKRTKVIYKTLTPQWNQTLEF--PDDGSP----LMLYVKDHNALLPTSSIGECVVEYQRL 102
K+ TK T +P++N+T +F ++ P L L VKD+ IG V++ Q +
Sbjct: 2090 KQGTKTKSNTWSPKYNETFQFILGNENRPGAYELHLSVKDYCFAREDRIIGMTVIQLQNI 2149
Query: 103 PPNQMADKWIPL 114
W PL
Sbjct: 2150 AEKGSYGAWYPL 2161
>U13C_HUMAN (Q8NB66) Unc-13 homolog C (Munc13-3) (Fragment)
Length = 1190
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/119 (27%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+ + I + ++ + L A D G+SDPYV V G K+RTK I+ L P W++ F
Sbjct: 190 GTSKWSAKITITVVSAQGLQAKDKTGSSDPYVTVQVGKNKRRTKTIFGNLNPVWDEKFYF 249
Query: 70 P-------------DDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
D+ + VK H +G+ +VE + L D W L+
Sbjct: 250 ECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTLSGEM--DVWYNLE 306
Score = 29.3 bits (64), Expect = 8.2
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 7/87 (8%)
Query: 49 KKRTKVIYKTLTPQWNQTLEF--PDDGSP----LMLYVKDHNALLPTSSIGECVVEYQRL 102
K+ TK T +P++N+T +F + P L L VKD+ IG V++ Q +
Sbjct: 1076 KQGTKTKSNTWSPKYNETFQFILGKENRPGAYELHLSVKDYCFAREDRIIGMTVIQLQNI 1135
Query: 103 PPNQMADKWIP-LQGVKRGEIHIQITR 128
W P L+ + E + I R
Sbjct: 1136 AEKGSYGAWYPLLKNISMDETGLTILR 1162
>RBF1_CAEEL (P41885) Rabphilin 1
Length = 1106
Score = 51.2 bits (121), Expect = 2e-06
Identities = 51/222 (22%), Positives = 97/222 (42%), Gaps = 24/222 (10%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHY--GNFKK---RTKVIYKTLTPQWNQTLEF--- 69
+++ LI ++L A D G SDPYV+ H GN K +K I KTL P+WN+ + +
Sbjct: 844 LKMHLIRAKNLKAMDSNGFSDPYVKFHLLPGNTKATKLTSKTIEKTLNPEWNEEMSYYGI 903
Query: 70 -PDDGSP--LMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQI 126
DD L + V D + + + +GE + ++L N+M K+ ++++
Sbjct: 904 TEDDKEKKILRVTVLDRDR-IGSDFLGETRIALKKLNDNEM----------KKFNLYLES 952
Query: 127 TRKVPEMQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLE--GLSTTLSELETL 184
VP+ K + + ++ Q Q + I +E ++ G S ++
Sbjct: 953 ALPVPQQTKEEENEDRGKINVGLQYNIQQGSLFININRCVELVGMDSTGFSDPYCKVSLT 1012
Query: 185 EDTQEGYVAQLETEQMLLLSKIKELGQEIINSSPSPSLSRRI 226
T + + A+ T++ L + E Q ++ P + +I
Sbjct: 1013 PITSKAHRAKTSTKKRTLNPEWNEQLQFVVPFKDLPKKTLQI 1054
>KPC1_DROME (P05130) Protein kinase C, brain isozyme (EC 2.7.1.-)
(PKC) (dPKC53E(BR))
Length = 679
Score = 51.2 bits (121), Expect = 2e-06
Identities = 53/202 (26%), Positives = 84/202 (41%), Gaps = 23/202 (11%)
Query: 24 EGRDLVAADLRGTSDPYVRVHY-----GNFKKRTKVIYKTLTPQWNQTLEF----PDDGS 74
EGR+L+ D G SDPYV+V KK+T+ I L P WN+TL + D
Sbjct: 197 EGRNLIPMDPNGLSDPYVKVKLIPDDKDQSKKKTRTIKACLNPVWNETLTYDLKPEDKDR 256
Query: 75 PLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQ 134
+++ V D + +G + N + W L GE + VP
Sbjct: 257 RILIEVWDWDRTSRNDFMGALSFGISEIIKNP-TNGWFKLLTQDEGEYY-----NVP--- 307
Query: 135 KRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGL--STTLSELETLEDTQEGYV 192
D E L KL Q P+Q K M+++ + + + + +T + ++ L G V
Sbjct: 308 ---CADDEQDLLKLKQKPSQKKPMVMRSDTNTHTSSKKDMIRATDFNFIKVLGKGSFGKV 364
Query: 193 AQLETEQMLLLSKIKELGQEII 214
E + L IK L +++I
Sbjct: 365 LLAERKGSEELYAIKILKKDVI 386
>UN13_CAEEL (P27715) Phorbol ester/diacylglycerol-binding protein
unc-13 (Uncoordinated protein 13)
Length = 2155
Score = 50.8 bits (120), Expect = 3e-06
Identities = 23/61 (37%), Positives = 34/61 (55%)
Query: 9 QGSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLE 68
+GS + I L ++ + L+A D G SDPYV G K+RT+ I++ L P WN+
Sbjct: 1150 EGSSKWSAKITLTVLCAQGLIAKDKTGKSDPYVTAQVGKTKRRTRTIHQELNPVWNEKFH 1209
Query: 69 F 69
F
Sbjct: 1210 F 1210
>U13C_MOUSE (Q8K0T7) Unc-13 homolog C (Munc13-3) (Fragment)
Length = 1430
Score = 50.8 bits (120), Expect = 3e-06
Identities = 33/119 (27%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+ + I + ++ + L A D G+SDPYV V G K+RTK I+ L P W++ F
Sbjct: 1210 GTSKWSAKITITVVSAQGLQAKDKTGSSDPYVTVQVGKNKRRTKTIFGNLNPVWDEKFFF 1269
Query: 70 P-------------DDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
D+ + VK H +G+ +VE + L D W L+
Sbjct: 1270 ECHNSTDRIKVRVWDEDDDIKSRVKQHFKKESDDFLGQTIVEVRTLSGEM--DVWYNLE 1326
>U13A_RAT (Q62768) Unc-13 homolog A (Munc13-1)
Length = 1735
Score = 50.8 bits (120), Expect = 3e-06
Identities = 24/60 (40%), Positives = 32/60 (53%)
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+ + I + ++ + L A D G+SDPYV V G KKRTK IY L P W + F
Sbjct: 683 GTSKWSAKISITVVCAQGLQAKDKTGSSDPYVTVQVGKTKKRTKTIYGNLNPVWEENFHF 742
>U13A_HUMAN (Q9UPW8) Unc-13 homolog A (Munc13-1) (Fragment)
Length = 1157
Score = 50.8 bits (120), Expect = 3e-06
Identities = 24/60 (40%), Positives = 32/60 (53%)
Query: 10 GSGSGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G+ + I + ++ + L A D G+SDPYV V G KKRTK IY L P W + F
Sbjct: 124 GTSKWSAKISITVVCAQGLQAKDKTGSSDPYVTVQVGKTKKRTKTIYGNLNPVWEENFHF 183
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,025,928
Number of Sequences: 164201
Number of extensions: 1104034
Number of successful extensions: 2905
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 79
Number of HSP's that attempted gapping in prelim test: 2623
Number of HSP's gapped (non-prelim): 262
length of query: 231
length of database: 59,974,054
effective HSP length: 107
effective length of query: 124
effective length of database: 42,404,547
effective search space: 5258163828
effective search space used: 5258163828
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146563.10


