

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.5 + phase: 2 /pseudo/partial
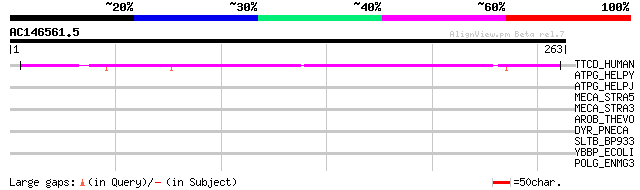
(263 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TTCD_HUMAN (Q8NBP0) Tetratricopeptide repeat protein 13 (TPR rep... 100 4e-21
ATPG_HELPY (P56082) ATP synthase gamma chain (EC 3.6.3.14) 32 1.2
ATPG_HELPJ (Q9ZK80) ATP synthase gamma chain (EC 3.6.3.14) 32 1.2
MECA_STRA5 (Q8E259) Adapter protein mecA 32 2.1
MECA_STRA3 (Q8E7L9) Adapter protein mecA 32 2.1
AROB_THEVO (Q978S6) 3-dehydroquinate synthase (EC 4.2.3.4) 32 2.1
DYR_PNECA (P16184) Dihydrofolate reductase (EC 1.5.1.3) 31 3.5
SLTB_BP933 (P09386) Shiga-like toxin II subunit B precursor (Ver... 30 6.0
YBBP_ECOLI (P77504) Hypothetical protein ybbP 30 7.8
POLG_ENMG3 (P32540) Genome polyprotein [Contains: Coat protein V... 30 7.8
>TTCD_HUMAN (Q8NBP0) Tetratricopeptide repeat protein 13 (TPR repeat
protein 13)
Length = 859
Score = 100 bits (249), Expect = 4e-21
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 596 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 651
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 652 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 711
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 712 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 770
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 771 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 828
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 829 TLPSVSETFPTLRSMIEVLNT 849
>ATPG_HELPY (P56082) ATP synthase gamma chain (EC 3.6.3.14)
Length = 301
Score = 32.3 bits (72), Expect = 1.2
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 4 NSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGK 59
N F + +L + + PNYER + K V+++ Y+ GKTD++I + N K
Sbjct: 131 NEYFSFNGIEVLDKINNLSSMPNYERVQEFMKKVVED--YLSGKTDKVIIIHNGFK 184
>ATPG_HELPJ (Q9ZK80) ATP synthase gamma chain (EC 3.6.3.14)
Length = 301
Score = 32.3 bits (72), Expect = 1.2
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 4 NSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGK 59
N F + +L + + PNYER + K V+++ Y+ GKTD++I + N K
Sbjct: 131 NEYFSFNGIEVLDKINNLSSMPNYERAQEFMKKVVED--YLSGKTDKVIIIHNGFK 184
>MECA_STRA5 (Q8E259) Adapter protein mecA
Length = 251
Score = 31.6 bits (70), Expect = 2.1
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPN--YERTLDIAKTVMKERSYVHGKTDQIIHLSNDG 58
E + +G G H ++L YF N Y R L+ A K R+Y+ + Q+IH
Sbjct: 184 ELYKNGKGYHMTILLDLENQPSYFANLMYARMLEHANVGTKTRAYLKEHSIQLIHDDAIS 243
Query: 59 KLEEI 63
KL+ I
Sbjct: 244 KLQMI 248
>MECA_STRA3 (Q8E7L9) Adapter protein mecA
Length = 251
Score = 31.6 bits (70), Expect = 2.1
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPN--YERTLDIAKTVMKERSYVHGKTDQIIHLSNDG 58
E + +G G H ++L YF N Y R L+ A K R+Y+ + Q+IH
Sbjct: 184 ELYKNGKGYHMTILLDLENQPSYFANLMYARMLEHANVGTKTRAYLKEHSIQLIHDDAIS 243
Query: 59 KLEEI 63
KL+ I
Sbjct: 244 KLQMI 248
>AROB_THEVO (Q978S6) 3-dehydroquinate synthase (EC 4.2.3.4)
Length = 360
Score = 31.6 bits (70), Expect = 2.1
Identities = 14/33 (42%), Positives = 22/33 (66%)
Query: 132 AMAWEALCNAYCGENYGSTDFDVLENVRDAILR 164
A+A L A+ GE YG+T+ +V+E +RD + R
Sbjct: 262 AVATGMLVEAHIGEKYGNTNHEVIEAIRDLMKR 294
>DYR_PNECA (P16184) Dihydrofolate reductase (EC 1.5.1.3)
Length = 206
Score = 30.8 bits (68), Expect = 3.5
Identities = 30/119 (25%), Positives = 46/119 (38%), Gaps = 11/119 (9%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
E + G G H+ L A + Y RT +V R +V G + KL
Sbjct: 84 ESLDLGNGIHSAKSLDHALELLY-----RTYGSESSVQINRIFVIGGAQLYKAAMDHPKL 138
Query: 61 EEIMHAKSCSDLYKVV------GEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDF 113
+ IM D++ V + WSS W + + GT+V K+ + GFD+
Sbjct: 139 DRIMATIIYKDIHCDVFFPLKFRDKEWSSVWKKEKHSDLESWVGTKVPHGKINEDGFDY 197
>SLTB_BP933 (P09386) Shiga-like toxin II subunit B precursor
(Verotoxin 2 subunit B) (SLT-IIB)
Length = 89
Score = 30.0 bits (66), Expect = 6.0
Identities = 14/46 (30%), Positives = 24/46 (51%)
Query: 58 GKLEEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTL 103
GK+E + + + KV G+++W+S W + QL G VT+
Sbjct: 25 GKIEFSKYNEDDTFTVKVDGKEYWTSRWNLQPLLQSAQLTGMTVTI 70
>YBBP_ECOLI (P77504) Hypothetical protein ybbP
Length = 804
Score = 29.6 bits (65), Expect = 7.8
Identities = 14/46 (30%), Positives = 23/46 (49%), Gaps = 1/46 (2%)
Query: 194 ANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLKVTTSW 239
A+++ TG++ G +V W + N +D + WL P LK W
Sbjct: 179 ADVDKTGAVQPGSRVTWRYKFGGNENQ-LDGYEKWLLPQLKPEQRW 223
>POLG_ENMG3 (P32540) Genome polyprotein [Contains: Coat protein VP4
(P1A); Coat protein VP2 (P1B); Coat protein VP3 (P1C);
Coat protein VP1 (P1D)] (Fragment)
Length = 901
Score = 29.6 bits (65), Expect = 7.8
Identities = 17/63 (26%), Positives = 26/63 (40%), Gaps = 1/63 (1%)
Query: 70 SDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTPARWEDYDA 129
+DL + D W + W T T ++ M G DF+++ P +PA W
Sbjct: 568 TDLVNITNADGWVTVW-QLTPLTYPPGCPTSAKILTMVSAGKDFSLKMPISPAPWSPQGV 626
Query: 130 EMA 132
E A
Sbjct: 627 ENA 629
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,252,626
Number of Sequences: 164201
Number of extensions: 1334452
Number of successful extensions: 2802
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2798
Number of HSP's gapped (non-prelim): 10
length of query: 263
length of database: 59,974,054
effective HSP length: 108
effective length of query: 155
effective length of database: 42,240,346
effective search space: 6547253630
effective search space used: 6547253630
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146561.5


