

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
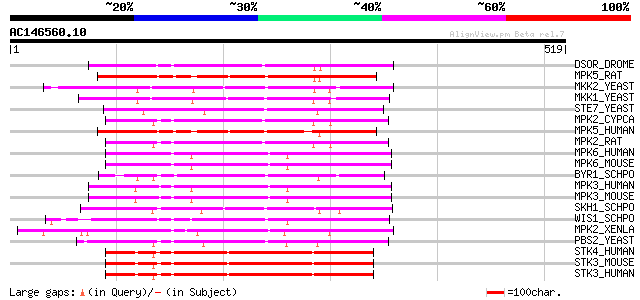
(519 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DSOR_DROME (Q24324) Dual specificity mitogen-activated protein k... 203 1e-51
MPK5_RAT (Q62862) Dual specificity mitogen-activated protein kin... 183 8e-46
MKK2_YEAST (P32491) MAP kinase kinase MKK2/SSP33 (EC 2.7.1.-) 182 1e-45
MKK1_YEAST (P32490) MAP kinase kinase MKK1/SSP32 (EC 2.7.1.-) 177 6e-44
STE7_YEAST (P06784) Serine/threonine-protein kinase STE7 (EC 2.7... 176 2e-43
MPK2_CYPCA (Q90321) Dual specificity mitogen-activated protein k... 174 6e-43
MPK5_HUMAN (Q13163) Dual specificity mitogen-activated protein k... 172 2e-42
MPK2_RAT (P36506) Dual specificity mitogen-activated protein kin... 172 2e-42
MPK6_HUMAN (P52564) Dual specificity mitogen-activated protein k... 169 1e-41
MPK6_MOUSE (P70236) Dual specificity mitogen-activated protein k... 169 2e-41
BYR1_SCHPO (P10506) Protein kinase byr1 (EC 2.7.1.-) (MAPK kinas... 168 3e-41
MPK3_HUMAN (P46734) Dual specificity mitogen-activated protein k... 168 4e-41
MPK3_MOUSE (O09110) Dual specificity mitogen-activated protein k... 166 1e-40
SKH1_SCHPO (Q9Y884) MAP kinase kinase skh1/pek1 (EC 2.7.1.-) 164 5e-40
WIS1_SCHPO (P33886) Protein kinase wis1 (EC 2.7.1.-) (Protein ki... 162 1e-39
MPK2_XENLA (Q07192) Dual specificity mitogen-activated protein k... 162 2e-39
PBS2_YEAST (P08018) Polymyxin B resistance protein kinase (EC 2.... 162 3e-39
STK4_HUMAN (Q13043) Serine/threonine-protein kinase 4 (EC 2.7.1.... 160 6e-39
STK3_MOUSE (Q9JI10) Serine/threonine-protein kinase 3 (EC 2.7.1.... 158 4e-38
STK3_HUMAN (Q13188) Serine/threonine-protein kinase 3 (EC 2.7.1.... 157 5e-38
>DSOR_DROME (Q24324) Dual specificity mitogen-activated protein
kinase kinase dSOR1 (EC 2.7.1.-) (Downstream of RAF)
(MAPKK)
Length = 393
Score = 203 bits (516), Expect = 1e-51
Identities = 125/312 (40%), Positives = 174/312 (55%), Gaps = 31/312 (9%)
Query: 74 EKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEK-RQQLLTEIRT 132
EK ++ G +GSG VV + H TH ++A K I++ K ++Q+L E++
Sbjct: 74 EKIGELSDEDLEKLGELGSGNGGVVMKVRHTHTHLIMARKLIHLEVKPAIKKQILRELKV 133
Query: 133 LCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLL 192
L E + +V F+GAFY+ G+ISI +EYMDGGSL IL+ IPE IL + +L
Sbjct: 134 LHECN-FPHIVGFYGAFYS--DGEISICMEYMDGGSLDLILKRAGRIPESILGRITLAVL 190
Query: 193 RGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSP 252
+GLSYL ++HRD+KP+N+LVN GE KI DFG+S L +S M +FVGT +YMSP
Sbjct: 191 KGLSYLRDNHAIIHRDVKPSNILVNSSGEIKICDFGVSGQLIDS--MANSFVGTRSYMSP 248
Query: 253 ERIRNESYSYPADIWSLGLALLESGTGEFPY--------------TANEGP--------- 289
ER++ YS +DIWSLGL+L+E G +P A E
Sbjct: 249 ERLQGTHYSVQSDIWSLGLSLVEMAIGMYPIPPPNTATLESIFADNAEESGQPTDEPRAM 308
Query: 290 --VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAK 347
L+ I+++P P + FS EF FVD CL+K PD R + LL HP+I K E +
Sbjct: 309 AIFELLDYIVNEPPPKLEHKIFSTEFKDFVDICLKKQPDERADLKTLLSHPWIRKAELEE 368
Query: 348 VDLPGFVRSVFD 359
VD+ G+V D
Sbjct: 369 VDISGWVCKTMD 380
>MPK5_RAT (Q62862) Dual specificity mitogen-activated protein kinase
kinase 5 (EC 2.7.1.-) (MAP kinase kinase 5) (MAPKK 5)
(MAPK/ERK kinase 5)
Length = 448
Score = 183 bits (465), Expect = 8e-46
Identities = 102/268 (38%), Positives = 164/268 (61%), Gaps = 17/268 (6%)
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEG 141
++R +G G V +A H+P+ +++A+K I + E ++Q+++E+ L +
Sbjct: 165 DIRYRDTLGHGNGGTVYKAYHVPSGKILAVKVILLDITLELQKQIMSELEILYKCDS-SY 223
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
++ F+GAF+ + +ISI E+MDGGSL ++R IPE +L + +++GL+YL +
Sbjct: 224 IIGFYGAFFVEN--RISICTEFMDGGSLD----VYRKIPEHVLGRIAVAVVKGLTYLWSL 277
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYS 261
+ L HRD+KP+N+LVN G+ K+ DFG+S L NS+A T+VGT YM+PERI E Y
Sbjct: 278 KIL-HRDVKPSNMLVNTSGQVKLCDFGVSTQLVNSIAK--TYVGTNAYMAPERISGEQYG 334
Query: 262 YPADIWSLGLALLESGTGEFPY---TANEG---PVNLMLQILDDPSPSPSKEKFSPEFCS 315
+D+WSLG++ +E G FPY N+G P+ L+ I+D+ SP +FS F
Sbjct: 335 IHSDVWSLGISFMELALGRFPYPQIQKNQGSLMPLQLLQCIVDEDSPVLPLGEFSEPFVH 394
Query: 316 FVDACLQKDPDNRPTAEQLLLHPFITKY 343
F+ C++K P RP E+L+ HPFI ++
Sbjct: 395 FITQCMRKQPKERPAPEELMGHPFIVQF 422
>MKK2_YEAST (P32491) MAP kinase kinase MKK2/SSP33 (EC 2.7.1.-)
Length = 506
Score = 182 bits (463), Expect = 1e-45
Identities = 123/346 (35%), Positives = 183/346 (52%), Gaps = 30/346 (8%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
+Y FS G S S + N + + VD+ ++ + E+ G +G
Sbjct: 167 AYDFSSSG-----SNSAPLSANNIISCSNLIQGKDVDQLEEEAWRFGHLKDEITTLGILG 221
Query: 92 SGASSVVQRAMHIPTHRVIALKKINIF--EKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
GA V + +V ALK IN + E ++Q+ E++ ++ + +V+++G F
Sbjct: 222 EGAGGSVAKCRLKNGKKVFALKTINTMNTDPEYQKQIFRELQ-FNKSFKSDYIVQYYGMF 280
Query: 150 YTPDSGQISIALEYMDGGSLA----DILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLV 205
S I IA+EYM G SL ++L+ I E ++ + + +LRGLSYLH R ++
Sbjct: 281 TDEQSSSIYIAMEYMGGKSLEATYKNLLKRGGRISERVIGKIAESVLRGLSYLHE-RKVI 339
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPAD 265
HRDIKP N+L+N KGE K+ DFG+S NS+AM TF GT YM+PERI+ + YS D
Sbjct: 340 HRDIKPQNILLNEKGEIKLCDFGVSGEAVNSLAM--TFTGTSFYMAPERIQGQPYSVTCD 397
Query: 266 IWSLGLALLESGTGEFPY-----TANEGPVNLMLQIL-------DDPSPSPSKEKFSPEF 313
+WSLGL LLE G FP+ T N P+ L+ IL D+P S +S F
Sbjct: 398 VWSLGLTLLEVAGGRFPFESDKITQNVAPIELLTMILTFSPQLKDEPELDIS---WSKTF 454
Query: 314 CSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFD 359
SF+D CL+KD RP+ Q+L HP+I KV++ FV+ ++
Sbjct: 455 RSFIDYCLKKDARERPSPRQMLKHPWIVGQMKKKVNMERFVKKCWE 500
>MKK1_YEAST (P32490) MAP kinase kinase MKK1/SSP32 (EC 2.7.1.-)
Length = 508
Score = 177 bits (449), Expect = 6e-44
Identities = 114/309 (36%), Positives = 168/309 (53%), Gaps = 25/309 (8%)
Query: 65 RSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIF--EKEK 122
R VD+ D+N Y + G +G GA V + ++ ALK IN + E
Sbjct: 202 RDVDQLDENGWKYANLKDRIETLGILGEGAGGSVSKCKLKNGSKIFALKVINTLNTDPEY 261
Query: 123 RQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSL----ADILRMHRT 178
++Q+ E++ + E +V ++G F ++ I IA+EYM G SL ++L
Sbjct: 262 QKQIFRELQ-FNRSFQSEYIVRYYGMFTDDENSSIYIAMEYMGGRSLDAIYKNLLERGGR 320
Query: 179 IPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVA 238
I E +L + + +LRGLSYLH + ++HRDIKP N+L+N G+ K+ DFG+S NS+A
Sbjct: 321 ISEKVLGKIAEAVLRGLSYLHEKK-VIHRDIKPQNILLNENGQVKLCDFGVSGEAVNSLA 379
Query: 239 MCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFP-----YTANEGPVNLM 293
TF GT YM+PERI+ + YS +D+WSLGL +LE G+FP AN P L+
Sbjct: 380 --TTFTGTSFYMAPERIQGQPYSVTSDVWSLGLTILEVANGKFPCSSEKMAANIAPFELL 437
Query: 294 LQIL-------DDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETA 346
+ IL D+P S +SP F SF+D CL+KD RP+ Q++ HP+I
Sbjct: 438 MWILTFTPELKDEPE---SNIIWSPSFKSFIDYCLKKDSRERPSPRQMINHPWIKGQMKK 494
Query: 347 KVDLPGFVR 355
V++ FVR
Sbjct: 495 NVNMEKFVR 503
>STE7_YEAST (P06784) Serine/threonine-protein kinase STE7 (EC
2.7.1.37)
Length = 515
Score = 176 bits (445), Expect = 2e-43
Identities = 104/283 (36%), Positives = 162/283 (56%), Gaps = 23/283 (8%)
Query: 88 GAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKR--QQLLTEIRTLCEAPCYEGLVEF 145
G IG+G S V +A+H+P +++A K I + + QL+ E+ + +E ++ F
Sbjct: 195 GKIGAGNSGTVVKALHVPDSKIVAKKTIPVEQNNSTIINQLVRELSIVKNVKPHENIITF 254
Query: 146 HGAFYTPD-SGQISIALEYMDGGSLADILRMHRTIP-------------EPILSSMFQKL 191
+GA+Y + +I I +EY D GSL IL +++ E +S + +
Sbjct: 255 YGAYYNQHINNEIIILMEYSDCGSLDKILSVYKRFVQRGTVSSKKTWFNELTISKIAYGV 314
Query: 192 LRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMS 251
L GL +L+ ++HRDIKP+N+L+N KG+ K+ DFG+S L NS+A TFVGT TYMS
Sbjct: 315 LNGLDHLYRQYKIIHRDIKPSNVLINSKGQIKLCDFGVSKKLINSIA--DTFVGTSTYMS 372
Query: 252 PERIRNESYSYPADIWSLGLALLESGTGEFPYTAN----EGPVNLMLQILDDPSPSPSKE 307
PERI+ YS D+WSLGL ++E TGEFP + +G ++L+ +I+++PSP K+
Sbjct: 373 PERIQGNVYSIKGDVWSLGLMIIELVTGEFPLGGHNDTPDGILDLLQRIVNEPSPRLPKD 432
Query: 308 K-FSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVD 349
+ +S E FV+ C K+ R + +LL H I KY + D
Sbjct: 433 RIYSKEMTDFVNRCCIKNERERSSIHELLHHDLIMKYVSPSKD 475
>MPK2_CYPCA (Q90321) Dual specificity mitogen-activated protein
kinase kinase 2 (EC 2.7.1.-) (MAP kinase kinase 2)
(MAPKK 2) (ERK activator kinase 2) (MAPK/ERK kinase 2)
(MEK2)
Length = 397
Score = 174 bits (440), Expect = 6e-43
Identities = 110/313 (35%), Positives = 166/313 (52%), Gaps = 55/313 (17%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEK-RQQLLTEIRTL--CEAPCYEGLVEFH 146
+G+G VV + H P+ V+A K I++ K R Q++ E++ L C +P +V F+
Sbjct: 75 LGAGNGGVVHKVRHKPSRLVMARKLIHLEIKPAIRNQIIRELQVLHECNSPY---IVGFY 131
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
GAFY+ G+ISI +E+MDGGSL +L+ R IPE IL + +LRGL YL ++H
Sbjct: 132 GAFYS--DGEISICMEHMDGGSLDQVLKEARRIPEEILGKVSIAVLRGLVYLREKHQIMH 189
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADI 266
RD+KP+N+LVN +GE K+ DFG+S L +S M +FVGT +YMSPER++ YS +D+
Sbjct: 190 RDVKPSNILVNSRGEIKLCDFGVSGQLIDS--MANSFVGTRSYMSPERLQGTHYSVQSDV 247
Query: 267 WSLGLALLESGTGEFP-----------------------------------------YTA 285
WS+GL+L+E G FP +
Sbjct: 248 WSMGLSLVELAIGRFPIPPPDAKELEAIFGRPVLDKGGAEGHSMSPRQRPPGRPVSGHGM 307
Query: 286 NEGPVNLMLQILD----DPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
+ P + ++LD +P P F+ +F FV CL K+P +R + L+ H FI
Sbjct: 308 DSRPAMAIFELLDYIVNEPPPKLPHGVFTTDFEEFVMKCLMKNPADRADLKMLMGHTFIK 367
Query: 342 KYETAKVDLPGFV 354
+ E +VD G++
Sbjct: 368 RAEVEEVDFAGWM 380
>MPK5_HUMAN (Q13163) Dual specificity mitogen-activated protein
kinase kinase 5 (EC 2.7.1.-) (MAP kinase kinase 5)
(MAPKK 5) (MAPK/ERK kinase 5)
Length = 438
Score = 172 bits (436), Expect = 2e-42
Identities = 97/265 (36%), Positives = 161/265 (60%), Gaps = 21/265 (7%)
Query: 83 EMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEG 141
++R +G G V +A H+P+ +++A+K I + E ++Q+++E+ L +
Sbjct: 165 DIRYRDTLGHGNGGTVYKAYHVPSGKILAVKVILLDITLELQKQIMSELEILYKCDS-SY 223
Query: 142 LVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGV 201
++ F+GAF+ + +ISI E+MDGGSL ++R +PE +L + +++GL+YL +
Sbjct: 224 IIGFYGAFFVEN--RISICTEFMDGGSLD----VYRKMPEHVLGRIAVAVVKGLTYLWSL 277
Query: 202 RYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYS 261
+ L HRD+KP+N+LVN +G+ K+ DFG+S L NS+A T+VGT YM+PERI E Y
Sbjct: 278 KIL-HRDVKPSNMLVNTRGQVKLCDFGVSTQLVNSIA--KTYVGTNAYMAPERISGEQYG 334
Query: 262 YPADIWSLGLALLESGTGEFPYTANEG---PVNLMLQILDDPSPSPSKEKFSPEFCSFVD 318
+D+WSLG++ +E N+G P+ L+ I+D+ SP +FS F F+
Sbjct: 335 IHSDVWSLGISFME-------IQKNQGSLMPLQLLQCIVDEDSPVLPVGEFSEPFVHFIT 387
Query: 319 ACLQKDPDNRPTAEQLLLHPFITKY 343
C++K P RP E+L+ HPFI ++
Sbjct: 388 QCMRKQPKERPAPEELMGHPFIVQF 412
>MPK2_RAT (P36506) Dual specificity mitogen-activated protein kinase
kinase 2 (EC 2.7.1.-) (MAP kinase kinase 2) (MAPKK 2)
(ERK activator kinase 2) (MAPK/ERK kinase 2) (MEK2)
Length = 400
Score = 172 bits (435), Expect = 2e-42
Identities = 110/313 (35%), Positives = 166/313 (52%), Gaps = 55/313 (17%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEK-RQQLLTEIRTL--CEAPCYEGLVEFH 146
+G+G VV +A H P+ ++A K I++ K R Q++ E++ L C +P +V F+
Sbjct: 78 LGAGNGGVVTKARHRPSGLIMARKLIHLEIKPAVRNQIIRELQVLHECNSPY---IVGFY 134
Query: 147 GAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
GAFY+ G+ISI +E+MDGGSL +L+ + IPE IL + +LRGL+YL ++H
Sbjct: 135 GAFYS--DGEISICMEHMDGGSLDQVLKEAKRIPEDILGKVSIAVLRGLAYLREKHQIMH 192
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADI 266
RD+KP+N+LVN +GE K+ DFG+S L +S M +FVGT +YMSPER++ YS +DI
Sbjct: 193 RDVKPSNILVNSRGEIKLCDFGVSGQLIDS--MANSFVGTRSYMSPERLQGTHYSVQSDI 250
Query: 267 WSLGLALLESGTGEFP-----------------------------------------YTA 285
WS+GL+L+E G +P +
Sbjct: 251 WSMGLSLVELAIGRYPIPPPDAKELEASFGRPVVDGADGEPHSVSPRPRPPGRPISGHGM 310
Query: 286 NEGPVNLMLQILD----DPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
+ P + ++LD +P P FS +F FV+ CL K+P R + L H FI
Sbjct: 311 DSRPAMAIFELLDYIVNEPPPKLPSGVFSSDFQEFVNKCLIKNPAERADLKLLTNHAFIK 370
Query: 342 KYETAKVDLPGFV 354
+ E VD G++
Sbjct: 371 RSEGEDVDFAGWL 383
>MPK6_HUMAN (P52564) Dual specificity mitogen-activated protein
kinase kinase 6 (EC 2.7.1.-) (MAP kinase kinase 6)
(MAPKK 6) (MAPK/ERK kinase 6) (SAPKK3)
Length = 334
Score = 169 bits (429), Expect = 1e-41
Identities = 95/276 (34%), Positives = 154/276 (55%), Gaps = 11/276 (3%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGA 148
+G GA VV++ H+P+ +++A+K+I +++++LL ++ V F+GA
Sbjct: 59 LGRGAYGVVEKMRHVPSGQIMAVKRIRATVNSQEQKRLLMDLDISMRTVDCPFTVTFYGA 118
Query: 149 FYTPDSGQISIALEYMDGGS---LADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLV 205
+ G + I +E MD ++ +TIPE IL + +++ L +LH ++
Sbjct: 119 LFR--EGDVWICMELMDTSLDKFYKQVIDKGQTIPEDILGKIAVSIVKALEHLHSKLSVI 176
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE----SYS 261
HRD+KP+N+L+N G+ K+ DFGIS L +SVA G YM+PERI E YS
Sbjct: 177 HRDVKPSNVLINALGQVKMCDFGISGYLVDSVAKTID-AGCKPYMAPERINPELNQKGYS 235
Query: 262 YPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACL 321
+DIWSLG+ ++E FPY + P + Q++++PSP +KFS EF F CL
Sbjct: 236 VKSDIWSLGITMIELAILRFPYDSWGTPFQQLKQVVEEPSPQLPADKFSAEFVDFTSQCL 295
Query: 322 QKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSV 357
+K+ RPT +L+ HPF T +E+ D+ FV+ +
Sbjct: 296 KKNSKERPTYPELMQHPFFTLHESKGTDVASFVKLI 331
>MPK6_MOUSE (P70236) Dual specificity mitogen-activated protein
kinase kinase 6 (EC 2.7.1.-) (MAP kinase kinase 6)
(MAPKK 6) (MAPK/ERK kinase 6) (SAPKK3)
Length = 334
Score = 169 bits (428), Expect = 2e-41
Identities = 94/276 (34%), Positives = 154/276 (55%), Gaps = 11/276 (3%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGA 148
+G GA VV++ H+P+ +++A+K+I +++++LL ++ V F+GA
Sbjct: 59 LGRGAYGVVEKMRHVPSGQIMAVKRIRATVNSQEQKRLLMDLDVSMRTVDCPFTVTFYGA 118
Query: 149 FYTPDSGQISIALEYMDGGS---LADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLV 205
+ G + I +E MD ++ +TIPE IL + +++ L +LH ++
Sbjct: 119 LFR--EGDVWICMELMDTSLDKFYKQVIDKGQTIPEDILGKIAVSIVKALEHLHSKLSVI 176
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE----SYS 261
HRD+KP+N+L+N G+ K+ DFGIS L +SVA G YM+PERI E YS
Sbjct: 177 HRDVKPSNVLINTLGQVKMCDFGISGYLVDSVAKTID-AGCKPYMAPERINPELNQKGYS 235
Query: 262 YPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACL 321
+DIWSLG+ ++E FPY + P + Q++++PSP +KFS +F F CL
Sbjct: 236 VKSDIWSLGITMIELAILRFPYDSWGTPFQQLKQVVEEPSPQLPADKFSADFVDFTSQCL 295
Query: 322 QKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSV 357
+K+ RPT +L+ HPF T +E+ D+ FV+ +
Sbjct: 296 KKNSKERPTYPELMQHPFFTVHESKAADVASFVKLI 331
>BYR1_SCHPO (P10506) Protein kinase byr1 (EC 2.7.1.-) (MAPK kinase)
(MAPKK)
Length = 340
Score = 168 bits (426), Expect = 3e-41
Identities = 109/278 (39%), Positives = 164/278 (58%), Gaps = 27/278 (9%)
Query: 84 MRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIF---EKEKRQQLLTEIRTL--CEAPC 138
+R G GA S+V+ HR I + + ++ + + ++Q+L E+ L C +P
Sbjct: 69 VRHLGEGNGGAVSLVK-------HRNIFMARKTVYVGSDSKLQKQILRELGVLHHCRSPY 121
Query: 139 YEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYL 198
+V F+GAF ++ IS+ +EYMD GSL ILR IP IL + +++GL YL
Sbjct: 122 ---IVGFYGAFQYKNN--ISLCMEYMDCGSLDAILREGGPIPLDILGKIINSMVKGLIYL 176
Query: 199 HGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE 258
+ V +++HRD+KP+N++VN +GE K+ DFG+S L NSVA TFVGT TYMSPERIR
Sbjct: 177 YNVLHIIHRDLKPSNVVVNSRGEIKLCDFGVSGELVNSVAQ--TFVGTSTYMSPERIRGG 234
Query: 259 SYSYPADIWSLGLALLESGTGEFPYTANE-----GPVNLMLQIL-DDPSPSPSKEKFSPE 312
Y+ +DIWSLG++++E T E P++ + G ++L+ I+ ++P PS F +
Sbjct: 235 KYTVKSDIWSLGISIIELATQELPWSFSNIDDSIGILDLLHCIVQEEPPRLPS--SFPED 292
Query: 313 FCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDL 350
FVDACL KDP R + +QL P+ + VDL
Sbjct: 293 LRLFVDACLHKDPTLRASPQQLCAMPYFQQALMINVDL 330
>MPK3_HUMAN (P46734) Dual specificity mitogen-activated protein
kinase kinase 3 (EC 2.7.1.-) (MAP kinase kinase 3)
(MAPKK 3) (MAPK/ERK kinase 3)
Length = 347
Score = 168 bits (425), Expect = 4e-41
Identities = 95/293 (32%), Positives = 161/293 (54%), Gaps = 13/293 (4%)
Query: 74 EKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKIN--IFEKEKRQQLLTEIR 131
++ + + ++ +G GA VV++ H + ++A+K+I + +E+++ L+
Sbjct: 54 DRNFEVEADDLVTISELGRGAYGVVEKVRHAQSGTIMAVKRIRATVNSQEQKRLLMDLDI 113
Query: 132 TLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGS---LADILRMHRTIPEPILSSMF 188
+ C+ V F+GA + G + I +E MD +L + TIPE IL +
Sbjct: 114 NMRTVDCFY-TVTFYGALFR--EGDVWICMELMDTSLDKFYRKVLDKNMTIPEDILGEIA 170
Query: 189 QKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVT 248
++R L +LH ++HRD+KP+N+L+N +G K+ DFGIS L +SVA G
Sbjct: 171 VSIVRALEHLHSKLSVIHRDVKPSNVLINKEGHVKMCDFGISGYLVDSVAKTMD-AGCKP 229
Query: 249 YMSPERIRNE----SYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSP 304
YM+PERI E Y+ +D+WSLG+ ++E FPY + P + Q++++PSP
Sbjct: 230 YMAPERINPELNQKGYNVKSDVWSLGITMIEMAILRFPYESWGTPFQQLKQVVEEPSPQL 289
Query: 305 SKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSV 357
++FSPEF F CL+K+P R + +L+ HPF T ++T K D+ FV+ +
Sbjct: 290 PADRFSPEFVDFTAQCLRKNPAERMSYLELMEHPFFTLHKTKKTDIAAFVKEI 342
>MPK3_MOUSE (O09110) Dual specificity mitogen-activated protein
kinase kinase 3 (EC 2.7.1.-) (MAP kinase kinase 3)
(MAPKK 3) (MAPK/ERK kinase 3)
Length = 347
Score = 166 bits (421), Expect = 1e-40
Identities = 94/293 (32%), Positives = 160/293 (54%), Gaps = 13/293 (4%)
Query: 74 EKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKIN--IFEKEKRQQLLTEIR 131
++ + + ++ +G GA VV++ H + ++A+K+I + +E+++ L+
Sbjct: 54 DRNFEVEADDLVTISELGRGAYGVVEKVRHAQSGTIMAVKRIRATVNTQEQKRLLMDLDI 113
Query: 132 TLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGS---LADILRMHRTIPEPILSSMF 188
+ C+ V F+GA + G + I +E MD +L + IPE IL +
Sbjct: 114 NMRTVDCFY-TVTFYGALFR--EGDVWICMELMDTSLDKFYRKVLEKNMKIPEDILGEIA 170
Query: 189 QKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVT 248
++R L +LH ++HRD+KP+N+L+N +G K+ DFGIS L +SVA G
Sbjct: 171 VSIVRALEHLHSKLSVIHRDVKPSNVLINKEGHVKMCDFGISGYLVDSVAKTMD-AGCKP 229
Query: 249 YMSPERIRNE----SYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSP 304
YM+PERI E Y+ +D+WSLG+ ++E FPY + P + Q++++PSP
Sbjct: 230 YMAPERINPELNQKGYNVKSDVWSLGITMIEMAILRFPYESWGTPFQQLKQVVEEPSPQL 289
Query: 305 SKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSV 357
++FSPEF F CL+K+P R + +L+ HPF T ++T K D+ FV+ +
Sbjct: 290 PADQFSPEFVDFTSQCLRKNPAERMSYLELMEHPFFTLHKTKKTDIAAFVKEI 342
>SKH1_SCHPO (Q9Y884) MAP kinase kinase skh1/pek1 (EC 2.7.1.-)
Length = 363
Score = 164 bits (415), Expect = 5e-40
Identities = 110/307 (35%), Positives = 166/307 (53%), Gaps = 22/307 (7%)
Query: 67 VDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQ-Q 125
++E DD+ + + ++G G S V++ T + A+K + Q Q
Sbjct: 62 LEELDDDHLHELVTNGGILYMNSLGEGVSGSVRKCRIRGTQMIFAMKTVLAAPNTALQKQ 121
Query: 126 LLTEIRT--LCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRT----I 179
LL E++ C +P +V+++GA Y Q++IA+EY GSL I + R+
Sbjct: 122 LLRELKINRSCTSPY---IVKYYGACYNNAECQLNIAMEYCGAGSLDAIYKRVRSQGGRT 178
Query: 180 PEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAM 239
E L + +L GLSYLH R ++HRDIKP+N+L+ KG+ K+ DFG+S L NS+A
Sbjct: 179 GERPLGKIAFGVLSGLSYLHD-RKIIHRDIKPSNILLTSKGQVKLCDFGVSGELVNSLA- 236
Query: 240 CATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEG-----PVNLML 294
TF GT YM+PERI SY+ +DIWSLGL L+E FP+ EG P+ L+
Sbjct: 237 -GTFTGTSYYMAPERISGGSYTISSDIWSLGLTLMEVALNRFPFPP-EGSPPPMPIELLS 294
Query: 295 QILDDPSPSPSKE---KFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLP 351
I++ P P +E K+S F F+ CL KD RP +++L HP++ +E VD+
Sbjct: 295 YIINMPPPLLPQEPGIKWSKSFQHFLCVCLDKDKTRRPGPQKMLTHPWVKAFERIHVDME 354
Query: 352 GFVRSVF 358
F+R V+
Sbjct: 355 EFLRQVW 361
>WIS1_SCHPO (P33886) Protein kinase wis1 (EC 2.7.1.-) (Protein
kinase sty2)
Length = 605
Score = 162 bits (411), Expect = 1e-39
Identities = 112/336 (33%), Positives = 173/336 (51%), Gaps = 35/336 (10%)
Query: 34 TFSD-----GGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFG 88
TFSD GT+N +++ +N G+ S S ++R E+
Sbjct: 280 TFSDILDAKSGTLNFKNKAV----LNSEGVNFSSGS-----------SFRINMSEIIKLE 324
Query: 89 AIGSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHG 147
+G G VV +A+H PT +ALK+I + E+ Q++ E+ L +A +V+F+G
Sbjct: 325 ELGKGNYGVVYKALHQPTGVTMALKEIRLSLEEATFNQIIMELDILHKAVS-PYIVDFYG 383
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHR 207
AF+ G + I +EYMD GS+ D L E +L+ +++GL L ++HR
Sbjct: 384 AFFV--EGSVFICMEYMDAGSM-DKLYAGGIKDEGVLARTAYAVVQGLKTLKEEHNIIHR 440
Query: 208 DIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNE-------SY 260
D+KP N+LVN G+ K+ DFG+S L S++ T +G +YM+PERIR +Y
Sbjct: 441 DVKPTNVLVNSNGQVKLCDFGVSGNLVASISK--TNIGCQSYMAPERIRVGGPTNGVLTY 498
Query: 261 SYPADIWSLGLALLESGTGEFPYTANE-GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDA 319
+ AD+WSLGL +LE G +PY + L + D P + FSPE FV+
Sbjct: 499 TVQADVWSLGLTILEMALGAYPYPPESYTSIFAQLSAICDGDPPSLPDSFSPEARDFVNK 558
Query: 320 CLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVR 355
CL K+P RP +L HP++ KY+ A VD+ + +
Sbjct: 559 CLNKNPSLRPDYHELANHPWLLKYQNADVDMASWAK 594
>MPK2_XENLA (Q07192) Dual specificity mitogen-activated protein
kinase kinase 2 (EC 2.7.1.-) (MAP kinase kinase 2)
(MAPKK 2) (MAPK-ERK kinase 2) (Fragment)
Length = 446
Score = 162 bits (410), Expect = 2e-39
Identities = 119/386 (30%), Positives = 187/386 (47%), Gaps = 38/386 (9%)
Query: 8 RKKLTPLFDAENGFSSTSSFDPN----DSYTFSDGGTVNLLSRSYGVYNINELGLQKCST 63
++K L A F ST+ F N ++ + L SY ++ G + ST
Sbjct: 52 KRKALKLNFANPAFKSTAKFTLNPTIQSTHVMHKLDAIRKLETSYQKQDLRTSGAKALST 111
Query: 64 SR-------------SVDESD----DNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPT 106
+ S++ S E+ + + +++ G IG GA V + H P+
Sbjct: 112 NEQATKNRLERLRTHSIESSGKLKLSPEQHWDFTAEDLKDLGEIGRGAYGSVNKMSHTPS 171
Query: 107 HRVIALKKINIFEKEKRQ-QLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMD 165
+++A+K+I EK Q QLL ++ + + +V+F+GA + G I +E M
Sbjct: 172 GQIMAVKRIRSTVDEKEQKQLLMDLDVVMRSSDCPYIVQFYGALFR--EGDCWICMELM- 228
Query: 166 GGSLADILR-----MHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKG 220
S + + IPE IL + ++ L++L ++HRDIKP+N+L++ G
Sbjct: 229 ATSFDKFYKYVYSFLDDVIPEEILGKITLATVKALNHLKENLKIIHRDIKPSNILLDTNG 288
Query: 221 EPKITDFGISAGLENSVAMCATFVGTVTYMSPERI----RNESYSYPADIWSLGLALLES 276
K+ DFGIS L +S+A G YM+PERI + Y +D+WSLG+ L E
Sbjct: 289 NIKLCDFGISGQLVDSIAKTRD-AGCRPYMAPERIDPSASRQGYDVRSDVWSLGITLYEL 347
Query: 277 GTGEFPYTANEGPVNLMLQIL--DDPSPSPSKEK-FSPEFCSFVDACLQKDPDNRPTAEQ 333
TG FPY + + Q++ D P S S+E+ FSP F SFV+ CL KD RP ++
Sbjct: 348 ATGRFPYPKWNSVFDQLTQVVKGDPPQLSNSEEREFSPSFTSFVNQCLTKDESKRPKYKE 407
Query: 334 LLLHPFITKYETAKVDLPGFVRSVFD 359
LL HPFI YE VD+ G+V + +
Sbjct: 408 LLKHPFILMYEERTVDVAGYVGKILE 433
>PBS2_YEAST (P08018) Polymyxin B resistance protein kinase (EC
2.7.1.-)
Length = 668
Score = 162 bits (409), Expect = 3e-39
Identities = 103/306 (33%), Positives = 166/306 (53%), Gaps = 23/306 (7%)
Query: 63 TSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINI-FEKE 121
+S+ +D S N + R E+ +G G V + +H PT+ ++A K++ + ++
Sbjct: 341 SSKGIDFS--NGSSSRITLDELEFLDELGHGNYGNVSKVLHKPTNVIMATKEVRLELDEA 398
Query: 122 KRQQLLTEIRTL--CEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTI 179
K +Q+L E+ L C +P +V+F+GAF+ G + + +EYMDGGSL I I
Sbjct: 399 KFRQILMELEVLHKCNSPY---IVDFYGAFFI--EGAVYMCMEYMDGGSLDKIYDESSEI 453
Query: 180 ---PEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNL-KGEPKITDFGISAGLEN 235
EP L+ + ++ GL L ++HRD+KP N+L + +G K+ DFG+S L
Sbjct: 454 GGIDEPQLAFIANAVIHGLKELKEQHNIIHRDVKPTNILCSANQGTVKLCDFGVSGNLVA 513
Query: 236 SVAMCATFVGTVTYMSPERIRN-----ESYSYPADIWSLGLALLESGTGEFPYTAN--EG 288
S+A T +G +YM+PERI++ +Y+ +DIWSLGL++LE G +PY +
Sbjct: 514 SLAK--TNIGCQSYMAPERIKSLNPDRATYTVQSDIWSLGLSILEMALGRYPYPPETYDN 571
Query: 289 PVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKV 348
+ + I+D P P +KFS + FV CLQK P+ RPT L HP++ KY V
Sbjct: 572 IFSQLSAIVDGPPPRLPSDKFSSDAQDFVSLCLQKIPERRPTYAALTEHPWLVKYRNQDV 631
Query: 349 DLPGFV 354
+ ++
Sbjct: 632 HMSEYI 637
>STK4_HUMAN (Q13043) Serine/threonine-protein kinase 4 (EC 2.7.1.37)
(STE20-like kinase MST1) (MST-1) (Mammalian STE20-like
protein kinase 1) (Serine/threonine-protein kinase
Krs-2)
Length = 487
Score = 160 bits (406), Expect = 6e-39
Identities = 91/255 (35%), Positives = 158/255 (61%), Gaps = 13/255 (5%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTL--CEAPCYEGLVEFHG 147
+G G+ V +A+H T +++A+K++ + + Q+++ EI + C++P +V+++G
Sbjct: 36 LGEGSYGSVYKAIHKETGQIVAIKQVPV--ESDLQEIIKEISIMQQCDSP---HVVKYYG 90
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRM-HRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
+++ + + I +EY GS++DI+R+ ++T+ E ++++ Q L+GL YLH +R +H
Sbjct: 91 SYFK--NTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILQSTLKGLEYLHFMRK-IH 147
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADI 266
RDIK N+L+N +G K+ DFG++ L +++A T +GT +M+PE I+ Y+ ADI
Sbjct: 148 RDIKAGNILLNTEGHAKLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADI 207
Query: 267 WSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSK-EKFSPEFCSFVDACLQKDP 325
WSLG+ +E G+ PY A+ P+ + I +P P+ K E +S F FV CL K P
Sbjct: 208 WSLGITAIEMAEGKPPY-ADIHPMRAIFMIPTNPPPTFRKPELWSDNFTDFVKQCLVKSP 266
Query: 326 DNRPTAEQLLLHPFI 340
+ R TA QLL HPF+
Sbjct: 267 EQRATATQLLQHPFV 281
>STK3_MOUSE (Q9JI10) Serine/threonine-protein kinase 3 (EC 2.7.1.37)
(STE20-like kinase MST2) (MST-2) (Mammalian STE20-like
protein kinase 2)
Length = 497
Score = 158 bits (399), Expect = 4e-38
Identities = 91/255 (35%), Positives = 159/255 (61%), Gaps = 13/255 (5%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTL--CEAPCYEGLVEFHG 147
+G G+ V +A+H + +V+A+K++ + + Q+++ EI + C++P +V+++G
Sbjct: 33 LGEGSYGSVFKAIHKESGQVVAIKQVPV--ESDLQEIIKEISIMQQCDSPY---VVKYYG 87
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRM-HRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
+++ + + I +EY GS++DI+R+ ++T+ E ++++ + L+GL YLH +R +H
Sbjct: 88 SYFK--NTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILKSTLKGLEYLHFMRK-IH 144
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADI 266
RDIK N+L+N +G K+ DFG++ L +++A T +GT +M+PE I+ Y+ ADI
Sbjct: 145 RDIKAGNILLNTEGHAKLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADI 204
Query: 267 WSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSK-EKFSPEFCSFVDACLQKDP 325
WSLG+ +E G+ PY A+ P+ + I +P P+ K E +S +F FV CL K P
Sbjct: 205 WSLGITSIEMAEGKPPY-ADIHPMRAIFMIPTNPPPTFRKPELWSDDFTDFVKKCLVKSP 263
Query: 326 DNRPTAEQLLLHPFI 340
+ R TA QLL HPFI
Sbjct: 264 EQRATATQLLQHPFI 278
>STK3_HUMAN (Q13188) Serine/threonine-protein kinase 3 (EC 2.7.1.37)
(STE20-like kinase MST2) (MST-2) (Mammalian STE20-like
protein kinase 2) (Serine/threonine-protein kinase
Krs-1)
Length = 491
Score = 157 bits (398), Expect = 5e-38
Identities = 91/255 (35%), Positives = 160/255 (62%), Gaps = 13/255 (5%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTL--CEAPCYEGLVEFHG 147
+G G+ V +A+H + +V+A+K++ + + Q+++ EI + C++P +V+++G
Sbjct: 33 LGEGSYGSVFKAIHKESGQVVAIKQVPV--ESDLQEIIKEISIMQQCDSPY---VVKYYG 87
Query: 148 AFYTPDSGQISIALEYMDGGSLADILRM-HRTIPEPILSSMFQKLLRGLSYLHGVRYLVH 206
+++ + + I +EY GS++DI+R+ ++T+ E ++++ + L+GL YLH +R +H
Sbjct: 88 SYFK--NTDLWIVMEYCGAGSVSDIIRLRNKTLIEDEIATILKSTLKGLEYLHFMRK-IH 144
Query: 207 RDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADI 266
RDIK N+L+N +G K+ DFG++ L +++A T +GT +M+PE I+ Y+ ADI
Sbjct: 145 RDIKAGNILLNTEGHAKLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADI 204
Query: 267 WSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSK-EKFSPEFCSFVDACLQKDP 325
WSLG+ +E G+ PY A+ P+ + I +P P+ K E +S +F FV CL K+P
Sbjct: 205 WSLGITSIEMAEGKPPY-ADIHPMRAIFMIPTNPPPTFRKPELWSDDFTDFVKKCLVKNP 263
Query: 326 DNRPTAEQLLLHPFI 340
+ R TA QLL HPFI
Sbjct: 264 EQRATATQLLQHPFI 278
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,713,601
Number of Sequences: 164201
Number of extensions: 2907090
Number of successful extensions: 10617
Number of sequences better than 10.0: 1667
Number of HSP's better than 10.0 without gapping: 1088
Number of HSP's successfully gapped in prelim test: 579
Number of HSP's that attempted gapping in prelim test: 6428
Number of HSP's gapped (non-prelim): 1984
length of query: 519
length of database: 59,974,054
effective HSP length: 115
effective length of query: 404
effective length of database: 41,090,939
effective search space: 16600739356
effective search space used: 16600739356
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146560.10


