

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.11 - phase: 0
(367 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
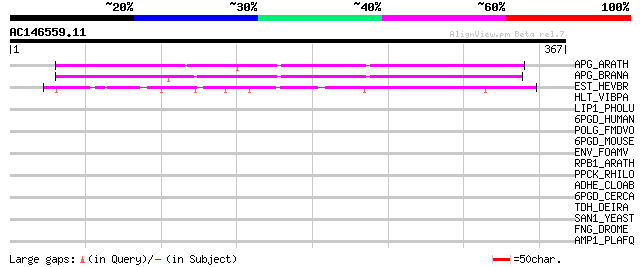
Score E
Sequences producing significant alignments: (bits) Value
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 187 4e-47
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 168 2e-41
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 93 9e-19
HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL) (Lecith... 40 0.007
LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3) (Triacylglyc... 35 0.39
6PGD_HUMAN (P52209) 6-phosphogluconate dehydrogenase, decarboxyl... 32 1.9
POLG_FMDVO (P03305) Genome polyprotein [Contains: Nonstructural ... 31 4.3
6PGD_MOUSE (Q9DCD0) 6-phosphogluconate dehydrogenase, decarboxyl... 31 4.3
ENV_FOAMV (P14351) Env polyprotein (Coat polyprotein) 31 5.7
RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subun... 30 7.4
PPCK_RHILO (Q98CL7) Phosphoenolpyruvate carboxykinase [ATP] (EC ... 30 7.4
ADHE_CLOAB (P33744) Aldehyde-alcohol dehydrogenase [Includes: Al... 30 7.4
6PGD_CERCA (P41570) 6-phosphogluconate dehydrogenase, decarboxyl... 30 7.4
TDH_DEIRA (Q9RTU4) L-threonine 3-dehydrogenase (EC 1.1.1.103) 30 9.7
SAN1_YEAST (P22470) SAN1 protein 30 9.7
FNG_DROME (Q24342) Fringe glycosyltransferase (EC 2.4.1.222) (O-... 30 9.7
AMP1_PLAFQ (O96935) M1-family aminopeptidase (EC 3.4.11.-) (Pfa-M1) 30 9.7
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 187 bits (475), Expect = 4e-47
Identities = 107/322 (33%), Positives = 171/322 (52%), Gaps = 17/322 (5%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A F FGDSV D GNNN L T +++ PYG+DF TGRFSNG+ D ++ +G++
Sbjct: 204 AVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAKYMGVKE 263
Query: 91 SLP-YLSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQI 149
+P YL P + LL G +FAS G G N T + I + QL F Y +K++ +
Sbjct: 264 IVPAYLDPKIQPNDLLTGVSFASGGAG-YNPTTSEAANAIPMLDQLTYFQDYIEKVNRLV 322
Query: 150 ----------GAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLIS 199
G E QL++K + +++ G ND + Y+ A+ + + +Y T +
Sbjct: 323 RQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYF--GSGAQRLKNDIDSYTTIIAD 380
Query: 200 EYKKILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMI 259
+ +LY GARR+ V GT P+GC P++ K + C+ EL A+ L+N +L+ ++
Sbjct: 381 SAASFVLQLYGYGARRIGVIGTPPLGCVPSQRLKKKK--ICNEELNYASQLFNSKLLLIL 438
Query: 260 TQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLC-TPISKLC 318
QL++ + + F+ ++ + + + P A+GF K CC G + LC SK+C
Sbjct: 439 GQLSKTLPNSTFVYMDIYTIISQMLETPAAYGFEETKKPCCKTGLLSAGALCKKSTSKIC 498
Query: 319 PNRNLYAFWDAFHPSEKASRII 340
PN + Y FWD HP+++A + I
Sbjct: 499 PNTSSYLFWDGVHPTQRAYKTI 520
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 168 bits (426), Expect = 2e-41
Identities = 101/317 (31%), Positives = 164/317 (50%), Gaps = 13/317 (4%)
Query: 31 AFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEP 90
A F FGDS+ D GNNN L T + + PYG+DFP TGRFSNG D S+ LG++
Sbjct: 125 AVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYISKYLGVKE 184
Query: 91 SLP-YLSPLLVGEK------LLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQ 143
+P Y+ L LL G +FAS G G L T + ++ + QL F Y++
Sbjct: 185 IVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTS-ESWKVTTMLDQLTYFQDYKK 243
Query: 144 KLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKK 203
++ +G + K++V+K +++ G ND + Y+ A+ + + ++ T +
Sbjct: 244 RMKKLVGKKKTKKIVSKGAAIVVAGSNDLIYTYF--GNGAQHLKNDVDSFTTMMADSAAS 301
Query: 204 ILQRLYDLGARRVLVTGTGPMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLN 263
+ +LY GARR+ V GT P+GC P++ K + C+ +L AA L+N +LV ++ QL+
Sbjct: 302 FVLQLYGYGARRIGVIGTPPIGCTPSQRVKKKK--ICNEDLNYAAQLFNSKLVIILGQLS 359
Query: 264 REIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPIS-KLCPNRN 322
+ + + + + + + + +P+ +GF K CC G G C + K N +
Sbjct: 360 KTLPNSTIVYGDIYSIFSKMLESPEDYGFEEIKKPCCKIGLTKGGVFCKERTLKNMSNAS 419
Query: 323 LYAFWDAFHPSEKASRI 339
Y FWD HPS++A I
Sbjct: 420 SYLFWDGLHPSQRAYEI 436
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 93.2 bits (230), Expect = 9e-19
Identities = 95/362 (26%), Positives = 146/362 (40%), Gaps = 53/362 (14%)
Query: 23 LAYAQPK---RAFFVFGDSVADNGNNNFLTTTARADAPPYGIDFPTHEPTGRFSNGLNIP 79
LAYA A F FGDS +D G PPYG F H TGR+S+G I
Sbjct: 23 LAYASETCDFPAIFNFGDSNSDTGGKAAAFYPLN---PPYGETF-FHRSTGRYSDGRLII 78
Query: 80 DLTSERLGLEPSLPYLSPLL--VGEKLLVGANFASAGVGILNDT-------GFQFLQIIH 130
D +E +LPYLSP L +G GA+FA+AG I T GF +
Sbjct: 79 DFIAESF----NLPYLSPYLSSLGSNFKHGADFATAGSTIKLPTTIIPAHGGFS---PFY 131
Query: 131 IGKQLDLFNQY--QQKLSAQIGAEGAKQL-----VNKAIVLIMLGGNDFVNNYYLVPFSA 183
+ Q F Q+ + + + G A+ + KA+ +G ND + + +
Sbjct: 132 LDVQYSQFRQFIPRSQFIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGF--LNLTV 189
Query: 184 RSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMGCAPAELAL----KSRNGD 239
++P+ V + KKI YDLGAR + TGP+GC L + +
Sbjct: 190 EEVNATVPDLVNSFSANVKKI----YDLGARTFWIHNTGPIGCLSFILTYFPWAEKDSAG 245
Query: 240 CDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDAC 299
C A +N +L +++ QL +++ F+ V+ + + + P+ GF C
Sbjct: 246 CAKAYNEVAQHFNHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFPLITC 305
Query: 300 CGQGRFNGIGLCTP-------------ISKLCPNRNLYAFWDAFHPSEKASRIIVQQMFI 346
CG G + P + C ++ WD H +E A+ Q+
Sbjct: 306 CGYGGKYNFSVTAPCGDTVTADDGTKIVVGSCACPSVRVNWDGAHYTEAANEYFFDQIST 365
Query: 347 GS 348
G+
Sbjct: 366 GA 367
>HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL)
(Lecithin-dependent haemolysin) (LDH) (Atypical
phospholipase) (Phospholipase A2) (Lysophospholipase)
Length = 418
Score = 40.4 bits (93), Expect = 0.007
Identities = 51/247 (20%), Positives = 97/247 (38%), Gaps = 36/247 (14%)
Query: 122 GFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKAIVLIMLGGNDFVNNYYLVPF 181
GF + + I K L L+N A GA G Q + + G + V++Y
Sbjct: 182 GFVWTEYIAKAKNLPLYNW------AVGGAAGENQYI------ALTGVGEQVSSYLTYAK 229
Query: 182 SARSR---------QFSLPNYVTY------LISEYKKILQRLYDLGARRVLVTGTGPMGC 226
A++ +F L +++ Y + ++Y + L RL D GA+ ++ T P
Sbjct: 230 LAKNYKPANTLFTLEFGLNDFMNYNRGVPEVKADYAEALIRLTDAGAKNFMLM-TLPDAT 288
Query: 227 APAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMDFITN 286
+ ++ + + +RA L + ++ + G ++ + + H + +
Sbjct: 289 KAPQFKYSTQE---EIDKIRAKVLEMNEFIKAQAMYYKAQGYNITL-FDTHALFETLTSA 344
Query: 287 PKAFGFVTAKDACCGQGRFNGIGLCTP--ISKLCP--NRNLYAFWDAFHPSEKASRIIVQ 342
P+ GFV A D C R + + + C + FWD HP+ R + +
Sbjct: 345 PEEHGFVNASDPCLDINRSSSVDYMYTHALRSECAASGAEKFVFWDVTHPTTATHRYVAE 404
Query: 343 QMFIGSN 349
+M SN
Sbjct: 405 KMLESSN 411
>LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3)
(Triacylglycerol lipase)
Length = 645
Score = 34.7 bits (78), Expect = 0.39
Identities = 25/81 (30%), Positives = 37/81 (44%), Gaps = 4/81 (4%)
Query: 262 LNREIGDDVFIAVNAHKMHMDFITNPKAFGFVTAKDACCGQGRFNGIGLCTPISKLCPNR 321
L++E G+ V + VNA + + I NP +GF+ C QG G C
Sbjct: 265 LSQENGNIVRVDVNA--LLHEVIANPLRYGFLNTIGYACAQG--VNAGSCRSKDTGFDAS 320
Query: 322 NLYAFWDAFHPSEKASRIIVQ 342
+ F D FHP+ +A I+ Q
Sbjct: 321 KPFLFADDFHPTPEAHHIVSQ 341
>6PGD_HUMAN (P52209) 6-phosphogluconate dehydrogenase,
decarboxylating (EC 1.1.1.44)
Length = 482
Score = 32.3 bits (72), Expect = 1.9
Identities = 31/116 (26%), Positives = 52/116 (44%), Gaps = 15/116 (12%)
Query: 118 LNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKA-----IVLIMLGG--- 169
+ND GF ++D F + K + +GA+ K++V+K I+L++ G
Sbjct: 21 MNDHGFVVCAFNRTVSKVDDFLANEAKGTKVVGAQSLKEMVSKLKKPRRIILLVKAGQAV 80
Query: 170 NDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMG 225
+DF+ LVP + SEY+ +R DL A+ +L G+G G
Sbjct: 81 DDFIEK--LVPLLDTGDII-----IDGGNSEYRDTTRRCRDLKAKGILFVGSGVSG 129
>POLG_FMDVO (P03305) Genome polyprotein [Contains: Nonstructural
protein P20A; Coat protein VP4; Coat protein VP2; Coat
protein VP3; Coat protein VP1; Core protein p12; Core
protein p34; Core protein p14; Genome-linked protein
VPG; Protease (EC 3.4.22.-);
Length = 2332
Score = 31.2 bits (69), Expect = 4.3
Identities = 20/80 (25%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Query: 42 NGNNNFLTTTARADAPPYGIDF--PTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLL 99
+G +TT + P YG F P ++ GRF+N L++ + L E +PY++
Sbjct: 513 DGYGGLVTTDPKTADPVYGKVFNPPRNQLPGRFTNLLDVAEACPTFLRFEGGVPYVTTKT 572
Query: 100 VGEKLLVGANFASAGVGILN 119
+++L + + A + N
Sbjct: 573 DSDRVLAQFDMSLAAKQMSN 592
>6PGD_MOUSE (Q9DCD0) 6-phosphogluconate dehydrogenase,
decarboxylating (EC 1.1.1.44)
Length = 482
Score = 31.2 bits (69), Expect = 4.3
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 15/116 (12%)
Query: 118 LNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKA-----IVLIMLGG--- 169
+ND GF ++D F + K + +GA+ K +V+K ++L++ G
Sbjct: 21 MNDHGFVVCAFNRTVSKVDDFLANEAKGTKVVGAQSLKDMVSKLKKPRRVILLVKAGQAV 80
Query: 170 NDFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMG 225
+DF+ LVP + SEY+ +R DL A+ +L G+G G
Sbjct: 81 DDFIEK--LVPLLDTGDII-----IDGGNSEYRDTTRRCRDLKAKGILFVGSGVSG 129
>ENV_FOAMV (P14351) Env polyprotein (Coat polyprotein)
Length = 985
Score = 30.8 bits (68), Expect = 5.7
Identities = 23/64 (35%), Positives = 28/64 (42%), Gaps = 4/64 (6%)
Query: 42 NGNNNFLTTTARADAPPYGIDFPT-HEPTGRFSNGLNIPDLTSERLGLEPSLPYLS---P 97
NG+ L ++ PPY T +E T F P + ERL EP LP L P
Sbjct: 805 NGSYLVLASSTDCQIPPYVPSIVTVNETTSCFGLDFKRPLVAEERLSFEPRLPNLQLRLP 864
Query: 98 LLVG 101
LVG
Sbjct: 865 HLVG 868
>RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6)
Length = 1840
Score = 30.4 bits (67), Expect = 7.4
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query: 56 APPYGIDFPTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGE 102
+P Y PT+ P+ +S+G + PD S G P+LP SP G+
Sbjct: 1771 SPSYSPSSPTYSPSSPYSSGAS-PDY-SPSAGYSPTLPGYSPSSTGQ 1815
>PPCK_RHILO (Q98CL7) Phosphoenolpyruvate carboxykinase [ATP] (EC
4.1.1.49) (PEP carboxykinase) (Phosphoenolpyruvate
carboxylase) (PEPCK)
Length = 536
Score = 30.4 bits (67), Expect = 7.4
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 4/72 (5%)
Query: 44 NNNFLTTTARADAPPYGIDF-PTHEPTGRFSNGLNIPDLTSERLGLEPSLPYLSPLLVGE 102
N+ LT R Y +DF P TGR S+ NI LT++ G+ P + L+P
Sbjct: 313 NDGRLTENTRC---AYPLDFIPNASKTGRASHPKNIIMLTADAFGVMPPIARLTPAQAMY 369
Query: 103 KLLVGANFASAG 114
L G AG
Sbjct: 370 HFLSGYTAKVAG 381
>ADHE_CLOAB (P33744) Aldehyde-alcohol dehydrogenase [Includes:
Alcohol dehydrogenase (EC 1.1.1.1) (ADH); Acetaldehyde
dehydrogenase [acetylating] (EC 1.2.1.10) (ACDH)]
Length = 862
Score = 30.4 bits (67), Expect = 7.4
Identities = 21/92 (22%), Positives = 40/92 (42%), Gaps = 2/92 (2%)
Query: 95 LSPLLVGEKLLVGANFASAGVGILNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGA 154
++P +VG+ A A AG+ + T ++ +G++ ++ + A A+
Sbjct: 290 VNPKIVGQSAYTIA--AMAGIKVPKTTRILIGEVTSLGEEEPFAHEKLSPVLAMYEADNF 347
Query: 155 KQLVNKAIVLIMLGGNDFVNNYYLVPFSARSR 186
+ KA+ LI LGG + Y AR +
Sbjct: 348 DDALKKAVTLINLGGLGHTSGIYADEIKARDK 379
>6PGD_CERCA (P41570) 6-phosphogluconate dehydrogenase,
decarboxylating (EC 1.1.1.44)
Length = 481
Score = 30.4 bits (67), Expect = 7.4
Identities = 30/116 (25%), Positives = 54/116 (45%), Gaps = 15/116 (12%)
Query: 118 LNDTGFQFLQIIHIGKQLDLFNQYQQKLSAQIGAEGAKQLVNKA-----IVLIMLGGN-- 170
+ND GF ++++ F + + K + IGA + +VNK I+L++ G+
Sbjct: 23 MNDKGFVVCAYNRTVEKVNQFLKNEAKGTNVIGATSLQDMVNKLKLPRKIMLLVKAGSAV 82
Query: 171 -DFVNNYYLVPFSARSRQFSLPNYVTYLISEYKKILQRLYDLGARRVLVTGTGPMG 225
DF+ LVP + + SEY+ +R +L A+++L G+G G
Sbjct: 83 DDFIQQ--LVPLLSPGDVI-----IDGGNSEYQDTARRCDELRAKKILYVGSGVSG 131
>TDH_DEIRA (Q9RTU4) L-threonine 3-dehydrogenase (EC 1.1.1.103)
Length = 348
Score = 30.0 bits (66), Expect = 9.7
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query: 176 YYLVP-FSARSRQFSLPNYVTYLISEYKKILQRL--YDLGARRVLVTGTGPMGCAPAELA 232
Y ++P F+A +P+ V + + + +DL VL+TG GP+GC A +A
Sbjct: 124 YLVLPAFNAFKLPDDIPDDVAAIFDPFGNAVHTALSFDLVGEDVLITGAGPIGCMAAAVA 183
>SAN1_YEAST (P22470) SAN1 protein
Length = 610
Score = 30.0 bits (66), Expect = 9.7
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 3/58 (5%)
Query: 16 NLFVGFDLAYAQPKRAFFVFGDSVADNGNNNFL---TTTARADAPPYGIDFPTHEPTG 70
N+ V +Y P+R + S DN NN +T A P +GI H+P G
Sbjct: 41 NITVSIQYSYFTPERLAHLSNISNNDNNENNSAASGSTIANGTGPSFGIGNGGHQPDG 98
>FNG_DROME (Q24342) Fringe glycosyltransferase (EC 2.4.1.222)
(O-fucosylpeptide
3-beta-N-acetylglucosaminyltransferase)
Length = 412
Score = 30.0 bits (66), Expect = 9.7
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 7/60 (11%)
Query: 223 PMGCAPAELALKSRNGDCDAELMRAASLYNPQLVQMITQLNREIGDDVFIAVNAHKMHMD 282
P ELA +SRNG+ +L + A PQ +T+L DD+FI+V K + D
Sbjct: 110 PTATLLTELARRSRNGELLRDLSQRAVTATPQ--PPVTEL-----DDIFISVKTTKNYHD 162
>AMP1_PLAFQ (O96935) M1-family aminopeptidase (EC 3.4.11.-) (Pfa-M1)
Length = 1085
Score = 30.0 bits (66), Expect = 9.7
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query: 149 IGAEGAKQLVNKAIVLIMLGGNDFV-NNYYLVPFSARSRQFSLPNYVTYLISEYKKILQR 207
I E K+++++ + + + FV NN + P + R FS P Y+ +++ ++IL
Sbjct: 696 INPENGKEMISQTTLELTKESDTFVFNNIAVKPIPSLFRGFSAPVYIEDNLTDEERILLL 755
Query: 208 LYDLGA 213
YD A
Sbjct: 756 KYDSDA 761
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,946,536
Number of Sequences: 164201
Number of extensions: 1807373
Number of successful extensions: 3713
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 3691
Number of HSP's gapped (non-prelim): 21
length of query: 367
length of database: 59,974,054
effective HSP length: 112
effective length of query: 255
effective length of database: 41,583,542
effective search space: 10603803210
effective search space used: 10603803210
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146559.11


