

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146555.15 - phase: 0 /pseudo
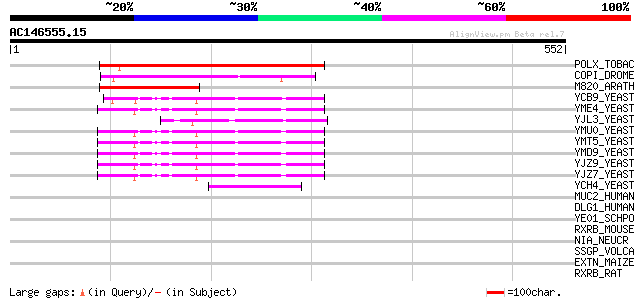
(552 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 161 4e-39
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 127 8e-29
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 84 7e-16
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 60 2e-08
YME4_YEAST (Q04711) Transposon Ty1 protein B 57 1e-07
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 57 1e-07
YMU0_YEAST (Q04670) Transposon Ty1 protein B 57 2e-07
YMT5_YEAST (Q04214) Transposon Ty1 protein B 57 2e-07
YMD9_YEAST (Q03434) Transposon Ty1 protein B 57 2e-07
YJZ9_YEAST (P47100) Transposon Ty1 protein B 57 2e-07
YJZ7_YEAST (P47098) Transposon Ty1 protein B 55 4e-07
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 50 2e-05
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 39 0.035
DLG1_HUMAN (Q12959) Presynaptic protein SAP97 (Synapse-associate... 35 0.51
YE01_SCHPO (O13798) Hypothetical protein C17H9.01 in chromosome I 35 0.66
RXRB_MOUSE (P28704) Retinoic acid receptor RXR-beta (Retinoid X ... 34 0.87
NIA_NEUCR (P08619) Nitrate reductase [NADPH] (EC 1.7.1.3) (NR) 33 1.5
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 33 1.9
EXTN_MAIZE (P14918) Extensin precursor (Proline-rich glycoprotein) 33 2.5
RXRB_RAT (P49743) Retinoic acid receptor RXR-beta (Retinoid X re... 32 3.3
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 161 bits (408), Expect = 4e-39
Identities = 91/229 (39%), Positives = 140/229 (60%), Gaps = 5/229 (2%)
Query: 90 EPKSYDEAIKHDCWKQVM---QNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDG 146
EP+S E + H Q+M Q E+ +L + GT+K+V+LP +P+ C WV K+K + D
Sbjct: 810 EPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDC 869
Query: 147 SIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLH 206
+ RYK LV KG+ Q +G+D+ + FS V K+T+IR +++LA+ + QLDV AFLH
Sbjct: 870 KLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLH 929
Query: 207 GDLHEDVYMAIPPGVSTS-KPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSN 265
GDL E++YM P G + K + VCKL+KSLYGLK A R+WY K + Y + S+
Sbjct: 930 GDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSD 989
Query: 266 ASLFTKK-NLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
++ K+ + ++FI+LL+YV D+ + G I +K L+++ K L
Sbjct: 990 PCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDL 1038
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 127 bits (319), Expect = 8e-29
Identities = 73/219 (33%), Positives = 121/219 (54%), Gaps = 6/219 (2%)
Query: 91 PKSYDEAIKHD---CWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGS 147
P S+DE D W++ + EL TW I P + + WV VK+N G+
Sbjct: 891 PNSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGN 950
Query: 148 IERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALASINY*FLHQLDVNNAFLHG 207
RYK LVA+G+ Q +DY +TF+ VA++++ R +++L +HQ+DV AFL+G
Sbjct: 951 PIRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNG 1010
Query: 208 DLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNAS 267
L E++YM +P G+S + N VCKL+K++YGLK A+R W+E + ++ +
Sbjct: 1011 TLKEEIYMRLPQGISCNSDN-VCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRC 1069
Query: 268 LF--TKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNAL 304
++ K N++ I +L+YV D+ +A ++ + K L
Sbjct: 1070 IYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYL 1108
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 84.3 bits (207), Expect = 7e-16
Identities = 42/99 (42%), Positives = 61/99 (61%)
Query: 90 EPKSYDEAIKHDCWKQVMQNELTTLDQTGTWKIVDLPPSSKPIGCIWVSKVKHNVDGSIE 149
EPKS A+K W Q MQ EL L + TW +V P + +GC WV K K + DG+++
Sbjct: 27 EPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLD 86
Query: 150 RYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLVIALA 188
R K LVAKG++Q EG+ + +T+S V + TIR ++ +A
Sbjct: 87 RLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 59.7 bits (143), Expect = 2e-08
Identities = 62/235 (26%), Positives = 111/235 (46%), Gaps = 28/235 (11%)
Query: 94 YDEAIKH-------DCWKQVMQNELTTLDQTGTWKIV------DLPPSSKPIGCIWVSKV 140
YDEAI + D + + E++ L + TW D+ P K I +++
Sbjct: 1249 YDEAITYNKDNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPK-KVINSMFIFNK 1307
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D+ V L+ +++A N ++ QL
Sbjct: 1308 KR--DGT---HKARFVARG--DIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQL 1360
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNG 258
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 1361 DISSAYLYADIKEELYIRPPPHLGLN--DKLLRLRKSLYGLKQSGANWYETIKSYLINCC 1418
Query: 259 YQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
Q S K +S + + ++V D+ L L+ I L + +KI+
Sbjct: 1419 DMQEVRGWSCVFK---NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKII 1470
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 57.0 bits (136), Expect = 1e-07
Identities = 59/236 (25%), Positives = 115/236 (48%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + + E+ L + TW ++ P + I +++
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPK-RVINSMFIFNR 865
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D+ V L+ ++LA N ++ QL
Sbjct: 866 KR--DGT---HKARFVARG--DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQL 918
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 919 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGANWYETIKSYLIKQC 976
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I L + +KI+
Sbjct: 977 GMEEVRGWSCVFK----NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKII 1028
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 57.0 bits (136), Expect = 1e-07
Identities = 46/170 (27%), Positives = 80/170 (47%), Gaps = 16/170 (9%)
Query: 151 YKVCLVAKGYNQIEGLDYFDTFSLVAKVTT----IRLVIALASINY*FLHQLDVNNAFLH 206
YK +V +G Q DT+S++ + I++ + +A+ F+ LD+N+AFL+
Sbjct: 1337 YKARIVCRGDTQSP-----DTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLY 1391
Query: 207 GDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNA 266
L E++Y+ P V KL+K+LYGLK + ++W + L G + +
Sbjct: 1392 AKLEEEIYIPHP-----HDRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTP 1446
Query: 267 SLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKILVSL 316
L+ + D +M+ VYV D +A + N L + KI +L
Sbjct: 1447 GLYQTE--DKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTL 1494
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 56.6 bits (135), Expect = 2e-07
Identities = 60/236 (25%), Positives = 114/236 (47%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + Q E+ L + TW ++ P + I +++
Sbjct: 807 YDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPK-RVINSMFIFNR 865
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D V L+ ++LA N ++ QL
Sbjct: 866 KR--DGT---HKARFVARG--DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQL 918
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 919 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGANWYETIKSYLIKQC 976
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I L + +KI+
Sbjct: 977 GMEEVRGWSCVFK----NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKII 1028
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 56.6 bits (135), Expect = 2e-07
Identities = 59/236 (25%), Positives = 115/236 (48%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + + E+ L + TW ++ P + I +++
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPK-RVINSMFIFNR 865
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D+ V L+ ++LA N ++ QL
Sbjct: 866 KR--DGT---HKARFVARG--DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQL 918
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 919 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGANWYETIKSYLIKQC 976
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I + L +KI+
Sbjct: 977 GMEEVRGWSCVFE----NSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKII 1028
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 56.6 bits (135), Expect = 2e-07
Identities = 59/236 (25%), Positives = 115/236 (48%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + + E+ L + TW ++ P + I +++
Sbjct: 807 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPK-RVINSMFIFNK 865
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D+ V L+ ++LA N ++ QL
Sbjct: 866 KR--DGT---HKARFVARG--DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQL 918
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 919 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGANWYETIKSYLIKQC 976
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I L + +KI+
Sbjct: 977 GMEEVRGWSCVFK----NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKII 1028
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 56.6 bits (135), Expect = 2e-07
Identities = 59/236 (25%), Positives = 114/236 (48%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + + E+ L + TW ++ P + I +++
Sbjct: 1234 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPK-RVINSMFIFNK 1292
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +D+ V L+ ++LA N ++ QL
Sbjct: 1293 KR--DGT---HKARFVARG--DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQL 1345
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KSLYGLK + WYE + I
Sbjct: 1346 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGANWYETIKSYLIQQC 1403
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I L +KI+
Sbjct: 1404 GMEEVRGWSCVFK----NSQVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKII 1455
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 55.5 bits (132), Expect = 4e-07
Identities = 59/236 (25%), Positives = 114/236 (48%), Gaps = 24/236 (10%)
Query: 88 YTEPKSYDEAIKH-DCWKQVMQNELTTLDQTGTWKI------VDLPPSSKPIGCIWVSKV 140
Y E +Y++ IK + + + E+ L + TW ++ P + I +++
Sbjct: 1234 YDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPK-RVINSMFIFNK 1292
Query: 141 KHNVDGSIERYKVCLVAKGYNQIEGLDYFDTFSLVAKVTTIRLV--IALASINY*FLHQL 198
K DG+ +K VA+G I+ D +DT V L+ ++LA N ++ QL
Sbjct: 1293 KR--DGT---HKARFVARG--DIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQL 1345
Query: 199 DVNNAFLHGDLHEDVYMAIPPGVSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPITN- 257
D+++A+L+ D+ E++Y+ PP + + +++ +L KS YGLK + WYE + I
Sbjct: 1346 DISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSHYGLKQSGANWYETIKSYLIKQC 1403
Query: 258 GYQQATSNASLFTKKNLDSFIMLLVYVYDITLAGDFLSEITFIKNALNQASKSKIL 313
G ++ + +F +S + + ++V D+ L L+ I L + +KI+
Sbjct: 1404 GMEEVRGWSCVFK----NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKII 1455
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 49.7 bits (117), Expect = 2e-05
Identities = 30/94 (31%), Positives = 48/94 (50%), Gaps = 1/94 (1%)
Query: 198 LDVNNAFLHGDLHEDVYMAIPPG-VSTSKPNQVCKLSKSLYGLKPASRKWYEKLTCLPIT 256
+DV+ AFL+ + E +Y+ PPG V+ P+ V +L +YGLK A W E +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 257 NGYQQATSNASLFTKKNLDSFIMLLVYVYDITLA 290
G+ + L+ + D I + VYV D+ +A
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVA 94
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 38.9 bits (89), Expect = 0.035
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 6/55 (10%)
Query: 2 TAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTT 56
T T +PTT+PSPPI+ T+ PP P TT + T T + PTT+TT
Sbjct: 1444 TTTTPLPTTTPSPPISTTTTPPPTTTPSPPTTTPSPPTTT------PSPPTTTTT 1492
Score = 37.4 bits (85), Expect = 0.10
Identities = 29/74 (39%), Positives = 35/74 (47%), Gaps = 10/74 (13%)
Query: 2 TAHTHIPTTSPSPPITKTSPSPPF---IPPPIRITTRNKITPTYM*DYICNIPTTSTTNN 58
T T PTT+PSPP T T+PSPP PP T + IT T + PTT+ T
Sbjct: 1647 TTTTPPPTTTPSPPTT-TTPSPPITTTTTPPPTTTPSSPITTT------PSPPTTTMTTP 1699
Query: 59 SHVQYPISNFLSHT 72
S P S + T
Sbjct: 1700 SPTTTPSSPITTTT 1713
Score = 36.6 bits (83), Expect = 0.17
Identities = 19/40 (47%), Positives = 21/40 (52%)
Query: 2 TAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
T T PTT+PSPP T T+ PP P ITT PT
Sbjct: 1412 TTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTPLPT 1451
Score = 35.8 bits (81), Expect = 0.30
Identities = 31/77 (40%), Positives = 37/77 (47%), Gaps = 13/77 (16%)
Query: 2 TAHTHIPTTSPSPPITKTSPSPPFI----PPPIRITTRNKITPTYM*DYICNIPTTSTTN 57
T T PTT+PSPP T T+PSPP I PPP TT + T T P +TT
Sbjct: 1568 TTTTPPPTTTPSPPTT-TTPSPPTITTTTPPP--TTTPSPPTTT------TTTPPPTTTP 1618
Query: 58 NSHVQYPISNFLSHTYL 74
+ PI+ S T L
Sbjct: 1619 SPPTTTPITPPTSTTTL 1635
Score = 35.0 bits (79), Expect = 0.51
Identities = 22/42 (52%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 2 TAHTHIPTTSPSPPI-TKTSPSPPFIP-PPIRITTRNKITPT 41
T T PTT+PSPPI T T+P P P PPI TT T T
Sbjct: 1428 TTTTPPPTTTPSPPITTTTTPLPTTTPSPPISTTTTPPPTTT 1469
Score = 33.5 bits (75), Expect = 1.5
Identities = 23/54 (42%), Positives = 28/54 (51%), Gaps = 7/54 (12%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITP-----TYM*DYICNIPTTSTT 56
PTT+PSPP T T+ PP P +TT ITP T + PTT+TT
Sbjct: 1480 PTTTPSPPTTTTTTPPPTTTPSPPMTT--PITPPASTTTLPPTTTPSPPTTTTT 1531
Score = 33.5 bits (75), Expect = 1.5
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query: 2 TAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
T T PTT+PSPP T+PSPP P TT PT
Sbjct: 1460 TTTTPPPTTTPSPP--TTTPSPPTTTPSPPTTTTTTPPPT 1497
Score = 33.5 bits (75), Expect = 1.5
Identities = 21/49 (42%), Positives = 25/49 (50%), Gaps = 12/49 (24%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTT 56
PTT+PSPP T T+ PP P TT ITP PT++TT
Sbjct: 1519 PTTTPSPPTTTTTTPPPTTTPSPPTTT--PITP----------PTSTTT 1555
Score = 32.7 bits (73), Expect = 2.5
Identities = 16/34 (47%), Positives = 18/34 (52%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
PTT+PSPP T T+ PP P TT PT
Sbjct: 1402 PTTTPSPPPTTTTTLPPTTTPSPPTTTTTTPPPT 1435
Score = 32.3 bits (72), Expect = 3.3
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query: 2 TAHTHIPTTSPSPPITKT----SPSPPFIPPPIRITTRNKITPTYM*DYICNIPTTSTTN 57
T T PTT+PS PIT T S + P PP T TP+ + +P T+T++
Sbjct: 1695 TMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSS 1754
Score = 31.6 bits (70), Expect = 5.6
Identities = 22/52 (42%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Query: 8 PTTSPS-PPITKTSPSPPFIPPPIRITTRN--KITPTYM*DYICNIPTTSTT 56
PTT+PS PP T T+P P P P TT + IT T P T+TT
Sbjct: 1558 PTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTT 1609
Score = 31.6 bits (70), Expect = 5.6
Identities = 20/52 (38%), Positives = 25/52 (47%), Gaps = 6/52 (11%)
Query: 8 PTTSPSPPITKTSPSPP----FIPPPIRITTRNKITPTYM*DYICNIPTTST 55
PTT+PSPP T+PSPP PPP + TP +P T+T
Sbjct: 1473 PTTTPSPP--TTTPSPPTTTTTTPPPTTTPSPPMTTPITPPASTTTLPPTTT 1522
Score = 30.8 bits (68), Expect = 9.6
Identities = 23/72 (31%), Positives = 29/72 (39%), Gaps = 18/72 (25%)
Query: 2 TAHTHIPTTSPSPPIT--------------KTSPSPP----FIPPPIRITTRNKITPTYM 43
T T PTT+PSPP+T T+PSPP PPP + TP
Sbjct: 1490 TTTTPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITP 1549
Query: 44 *DYICNIPTTST 55
+P T+T
Sbjct: 1550 PTSTTTLPPTTT 1561
>DLG1_HUMAN (Q12959) Presynaptic protein SAP97 (Synapse-associated
protein 97) (Discs, large homolog 1) (hDlg)
Length = 904
Score = 35.0 bits (79), Expect = 0.51
Identities = 33/141 (23%), Positives = 54/141 (37%), Gaps = 27/141 (19%)
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIP--------------------PPIRITTRNKITP 40
F +H+HI P+ + + P+ P IP PP+ + T + TP
Sbjct: 151 FVSHSHISPIKPTEAVLPSPPTVPVIPVLPVPAENTVILPTIPQANPPPVLVNTDSLETP 210
Query: 41 TYM----*DYICNIPTTSTTNNSHVQYPISNFLSHTYLSKSHSIFAMSLVSYTEPKSYDE 96
TY+ DY T NS + + I+ + ++ SIF +++
Sbjct: 211 TYVNGTDADYEYE-EITLERGNSGLGFSIAGGTDNPHIGDDSSIFITKIITGGAAAQDGR 269
Query: 97 AIKHDCWKQVMQNELTTLDQT 117
+DC QV NE+ D T
Sbjct: 270 LRVNDCILQV--NEVDVRDVT 288
>YE01_SCHPO (O13798) Hypothetical protein C17H9.01 in chromosome I
Length = 1202
Score = 34.7 bits (78), Expect = 0.66
Identities = 28/112 (25%), Positives = 52/112 (46%), Gaps = 10/112 (8%)
Query: 227 NQVCKLSKSLYGLKPASRKWYEKLTCLPITNGYQQATSNASLFTKKNLDSFIMLLVYVYD 286
N++ KL K + S EKLT +PIT +++ SN +L +KN+ L ++D
Sbjct: 316 NRISKLGKMI-----TSNHAEEKLTTIPITEAKKESVSNTALIARKNIIRERKYLSRMFD 370
Query: 287 ITLAGDFLSEITFIKNALNQASKSKILVSLNIS*ALKFVIPKVVFLYIIESI 338
I F ++ + +N +L L ++ K + + + L IE++
Sbjct: 371 I-----FEHIDSYFNHEINMLLVDLLLEPLQVAFVYKNLARRWISLEEIENL 417
>RXRB_MOUSE (P28704) Retinoic acid receptor RXR-beta (Retinoid X
receptor beta) (MHC class I regulatory element binding
protein H-2RIIBP)
Length = 520
Score = 34.3 bits (77), Expect = 0.87
Identities = 11/23 (47%), Positives = 17/23 (73%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPI 30
P++ P PP+T ++P PP PPP+
Sbjct: 94 PSSPPGPPLTPSAPPPPMPPPPL 116
>NIA_NEUCR (P08619) Nitrate reductase [NADPH] (EC 1.7.1.3) (NR)
Length = 982
Score = 33.5 bits (75), Expect = 1.5
Identities = 25/81 (30%), Positives = 41/81 (49%), Gaps = 10/81 (12%)
Query: 2 TAHTHIPTTSPSPPITKTS--PSPPFIPPPIRITTRNKITPTYM--*DYICNIPTTSTTN 57
+++T+ T+ P PIT +S P+ P PP R+TT I PT + D++ P
Sbjct: 92 SSNTNTNTSCPPSPITSSSLKPAYPLPPPSTRLTT---ILPTDLKTPDHLIRDPRLIRLT 148
Query: 58 NSH---VQYPISNFLSHTYLS 75
SH V+ P++ H +L+
Sbjct: 149 GSHPFNVEPPLTALFEHGFLT 169
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 33.1 bits (74), Expect = 1.9
Identities = 14/22 (63%), Positives = 14/22 (63%)
Query: 8 PTTSPSPPITKTSPSPPFIPPP 29
P SPSPP SPSPP PPP
Sbjct: 293 PPPSPSPPRKPPSPSPPVPPPP 314
>EXTN_MAIZE (P14918) Extensin precursor (Proline-rich glycoprotein)
Length = 267
Score = 32.7 bits (73), Expect = 2.5
Identities = 16/41 (39%), Positives = 18/41 (43%)
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
+T PT P+PP SP PP PP T TPT
Sbjct: 199 YTPSPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPT 239
Score = 31.2 bits (69), Expect = 7.3
Identities = 15/41 (36%), Positives = 18/41 (43%)
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
+T PT P+PP SP PP PP T TP+
Sbjct: 95 YTPSPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPS 135
Score = 31.2 bits (69), Expect = 7.3
Identities = 15/41 (36%), Positives = 18/41 (43%)
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
+T PT P+PP SP PP PP T TP+
Sbjct: 58 YTPSPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPS 98
Score = 30.8 bits (68), Expect = 9.6
Identities = 15/41 (36%), Positives = 18/41 (43%)
Query: 1 FTAHTHIPTTSPSPPITKTSPSPPFIPPPIRITTRNKITPT 41
+T PT P+PP SP PP PP T TP+
Sbjct: 21 YTPSPKPPTPKPTPPTYTPSPKPPASKPPTPKPTPPTYTPS 61
>RXRB_RAT (P49743) Retinoic acid receptor RXR-beta (Retinoid X
receptor beta) (Nuclear receptor coregulator 1)
(Fragment)
Length = 458
Score = 32.3 bits (72), Expect = 3.3
Identities = 11/23 (47%), Positives = 16/23 (68%)
Query: 8 PTTSPSPPITKTSPSPPFIPPPI 30
P++ P PP T ++P PP PPP+
Sbjct: 32 PSSPPGPPHTPSAPPPPMPPPPL 54
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.336 0.146 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,597,135
Number of Sequences: 164201
Number of extensions: 2282685
Number of successful extensions: 12919
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 12358
Number of HSP's gapped (non-prelim): 350
length of query: 552
length of database: 59,974,054
effective HSP length: 115
effective length of query: 437
effective length of database: 41,090,939
effective search space: 17956740343
effective search space used: 17956740343
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146555.15


