

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146554.5 + phase: 0
(236 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
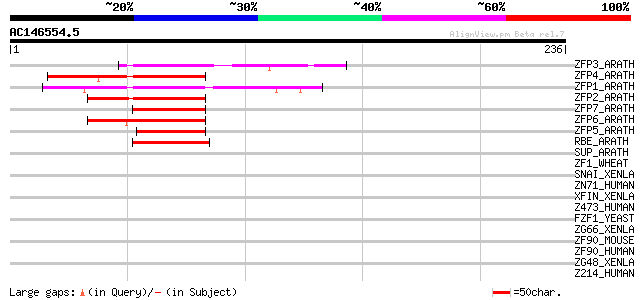
Score E
Sequences producing significant alignments: (bits) Value
ZFP3_ARATH (Q39262) Zinc finger protein 3 66 6e-11
ZFP4_ARATH (Q39263) Zinc finger protein 4 63 7e-10
ZFP1_ARATH (Q42485) Zinc finger protein 1 62 9e-10
ZFP2_ARATH (Q39261) Zinc finger protein 2 59 1e-08
ZFP7_ARATH (Q39266) Zinc finger protein 7 57 3e-08
ZFP6_ARATH (Q39265) Zinc finger protein 6 54 4e-07
ZFP5_ARATH (Q39264) Zinc finger protein 5 52 1e-06
RBE_ARATH (Q9LHS9) Probable transcriptional regulator RABBIT EARS 44 3e-04
SUP_ARATH (Q38895) Transcriptional regulator SUPERMAN 42 0.001
ZF1_WHEAT (Q42430) Zinc-finger protein 1 (WZF1) 38 0.018
SNAI_XENLA (P19382) Snail protein homolog Sna (xSna protein) 38 0.024
ZN71_HUMAN (Q9NQZ8) Endothelial zinc finger protein induced by t... 37 0.031
XFIN_XENLA (P08045) Zinc finger protein Xfin 36 0.070
Z473_HUMAN (Q8WTR7) Zinc finger protein 473 (Zinc finger protein... 36 0.091
FZF1_YEAST (P32805) Zinc finger protein FZF1 (Sulphite resistanc... 36 0.091
ZG66_XENLA (P18733) Gastrula zinc finger protein XLCGF66.1 (Frag... 35 0.12
ZF90_MOUSE (Q61967) Zinc finger protein 90 (Zfp-90) (Zinc finger... 35 0.12
ZF90_HUMAN (Q8TF47) Zinc finger protein 90 homolog (Zfp-90) (Fra... 35 0.12
ZG48_XENLA (P18723) Gastrula zinc finger protein XLCGF48.2 (Frag... 35 0.15
Z214_HUMAN (Q9UL59) Zinc finger protein 214 (BWSCR2 associated z... 35 0.15
>ZFP3_ARATH (Q39262) Zinc finger protein 3
Length = 235
Score = 66.2 bits (160), Expect = 6e-11
Identities = 43/100 (43%), Positives = 53/100 (53%), Gaps = 14/100 (14%)
Query: 47 EQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQ 106
EQK+ FSCNYC + F +SQALGGHQNAHKRER L K R + S+F H
Sbjct: 57 EQKL--FSCNYCQRTFYSSQALGGHQNAHKRERTLAKRGQR-------MAASASAFGHPY 107
Query: 107 GHS---YFRSANLHNPIGVHVTNAFPSWIGSPYGGYGGVY 143
G S + N H +G+ + S S Y G+GG Y
Sbjct: 108 GFSPLPFHGQYNNHRSLGIQAHSI--SHKLSSYNGFGGHY 145
>ZFP4_ARATH (Q39263) Zinc finger protein 4
Length = 260
Score = 62.8 bits (151), Expect = 7e-10
Identities = 34/68 (50%), Positives = 43/68 (63%), Gaps = 3/68 (4%)
Query: 17 LGTHTKSTSKPLENGGHSSS-FQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAH 75
L + S + PL + SS+ Q +P ++V FSCNYC +KF +SQALGGHQNAH
Sbjct: 50 LDLESSSLTLPLSSTSESSNPEQQQQQQPSVSKRV--FSCNYCQRKFYSSQALGGHQNAH 107
Query: 76 KRERVLKK 83
KRER L K
Sbjct: 108 KRERTLAK 115
>ZFP1_ARATH (Q42485) Zinc finger protein 1
Length = 228
Score = 62.4 bits (150), Expect = 9e-10
Identities = 50/135 (37%), Positives = 67/135 (49%), Gaps = 21/135 (15%)
Query: 15 LRLGTHTKSTSKPLEN---GGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGH 71
L GT T S +++ S+S E L + +V FSCNYC +KF +SQALGGH
Sbjct: 29 LSRGTATSSELNLIDSFKTSSSSTSHHQHQQEQLADPRV--FSCNYCQRKFYSSQALGGH 86
Query: 72 QNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYFR-----------SANLHNPI 120
QNAHKRER L K R + +M L+ SS + S R S N + +
Sbjct: 87 QNAHKRERTLAK---RGQYYKMTLSSLPSSAFAFGHGSVSRFASMASLPLHGSVNNRSTL 143
Query: 121 GV--HVTNAFPSWIG 133
G+ H T PS++G
Sbjct: 144 GIQAHSTIHKPSFLG 158
>ZFP2_ARATH (Q39261) Zinc finger protein 2
Length = 150
Score = 58.5 bits (140), Expect = 1e-08
Identities = 31/50 (62%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query: 34 SSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
SSS +T EQ V FSCNYC +KF +SQALGGHQNAHK ER L K
Sbjct: 34 SSSSSSTNSSSCLEQPRV-FSCNYCQRKFYSSQALGGHQNAHKLERTLAK 82
>ZFP7_ARATH (Q39266) Zinc finger protein 7
Length = 209
Score = 57.4 bits (137), Expect = 3e-08
Identities = 24/31 (77%), Positives = 27/31 (86%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
FSCNYC +KF +SQALGGHQNAHKRER + K
Sbjct: 59 FSCNYCRRKFYSSQALGGHQNAHKRERTMAK 89
>ZFP6_ARATH (Q39265) Zinc finger protein 6
Length = 197
Score = 53.5 bits (127), Expect = 4e-07
Identities = 26/54 (48%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query: 34 SSSFQNTLDEPLPEQ----KVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
++S QN EP P + ++ C YC ++F+ SQALGGHQNAHK+ER L K
Sbjct: 18 TTSVQNQSSEPRPGSGSGSESRKYECQYCCREFANSQALGGHQNAHKKERQLLK 71
>ZFP5_ARATH (Q39264) Zinc finger protein 5
Length = 211
Score = 52.0 bits (123), Expect = 1e-06
Identities = 21/29 (72%), Positives = 25/29 (85%)
Query: 55 CNYCDKKFSTSQALGGHQNAHKRERVLKK 83
C YC K+F+ SQALGGHQNAHK+ER+ KK
Sbjct: 62 CQYCGKEFANSQALGGHQNAHKKERLKKK 90
>RBE_ARATH (Q9LHS9) Probable transcriptional regulator RABBIT EARS
Length = 226
Score = 44.3 bits (103), Expect = 3e-04
Identities = 15/33 (45%), Positives = 25/33 (75%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKME 85
+SC++C ++F ++QALGGH N H+R+R K +
Sbjct: 55 YSCSFCGREFKSAQALGGHMNVHRRDRARLKQQ 87
>SUP_ARATH (Q38895) Transcriptional regulator SUPERMAN
Length = 204
Score = 42.4 bits (98), Expect = 0.001
Identities = 13/33 (39%), Positives = 26/33 (78%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKME 85
++C++C ++F ++QALGGH N H+R+R +++
Sbjct: 47 YTCSFCKREFRSAQALGGHMNVHRRDRARLRLQ 79
>ZF1_WHEAT (Q42430) Zinc-finger protein 1 (WZF1)
Length = 261
Score = 38.1 bits (87), Expect = 0.018
Identities = 14/25 (56%), Positives = 19/25 (76%)
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHK 76
EF C+ C K FS+ QALGGH+ +H+
Sbjct: 89 EFKCSVCGKSFSSYQALGGHKTSHR 113
Score = 33.5 bits (75), Expect = 0.45
Identities = 12/21 (57%), Positives = 15/21 (71%)
Query: 55 CNYCDKKFSTSQALGGHQNAH 75
C+ C K+F T QALGGH+ H
Sbjct: 162 CSICQKEFPTGQALGGHKRKH 182
>SNAI_XENLA (P19382) Snail protein homolog Sna (xSna protein)
Length = 259
Score = 37.7 bits (86), Expect = 0.024
Identities = 24/74 (32%), Positives = 32/74 (42%), Gaps = 5/74 (6%)
Query: 24 TSKPLENGGHSSSFQN--TLDEPLPEQKVVE---FSCNYCDKKFSTSQALGGHQNAHKRE 78
+SKPL+ SS + T D P P E F CN C K +ST L H+ H
Sbjct: 85 SSKPLDLTSFSSEDEGGKTSDPPSPASSATEAEKFQCNLCSKSYSTFAGLSKHKQLHCDS 144
Query: 79 RVLKKMEDRRREEE 92
+ K + E+E
Sbjct: 145 QTRKSFSCKYCEKE 158
>ZN71_HUMAN (Q9NQZ8) Endothelial zinc finger protein induced by
tumor necrosis factor alpha (Zinc finger protein 71)
(ZNF47)
Length = 489
Score = 37.4 bits (85), Expect = 0.031
Identities = 18/58 (31%), Positives = 28/58 (48%)
Query: 22 KSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRER 79
K P+ G + SS + P+P ++ + C+ C K FS S +L HQ H E+
Sbjct: 99 KGACPPVRRGKNFSSTSDLSKPPMPCEEKKTYDCSECGKAFSRSSSLIKHQRIHTGEK 156
>XFIN_XENLA (P08045) Zinc finger protein Xfin
Length = 1350
Score = 36.2 bits (82), Expect = 0.070
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHS 109
F C++CDKKF+ AL HQ H E+ K + + + T R + +H + H+
Sbjct: 410 FKCSHCDKKFTERSALAKHQRTHTGEKPYKCSDCGK-----EFTQRSNLILHQRIHT 461
Score = 32.0 bits (71), Expect = 1.3
Identities = 22/71 (30%), Positives = 32/71 (44%), Gaps = 8/71 (11%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKME-DRRREEEMDLTLRFSSFIHYQGHSYF 111
+ C C K+F+ S AL H+ H R + E R DLT H + H+ F
Sbjct: 248 YECPLCVKRFAESSALMKHKRTHSTHRPFRCSECSRSFTHNSDLT------AHMRKHTEF 301
Query: 112 RSA-NLHNPIG 121
R+ NL + +G
Sbjct: 302 RNVLNLDSVVG 312
Score = 30.8 bits (68), Expect = 2.9
Identities = 17/54 (31%), Positives = 26/54 (47%), Gaps = 9/54 (16%)
Query: 35 SSFQNTLD-------EPLPEQKVVE--FSCNYCDKKFSTSQALGGHQNAHKRER 79
+ F+N L+ +PL Q V +SC+ C K F ++ HQ H RE+
Sbjct: 299 TEFRNVLNLDSVVGTDPLSSQNVASSPYSCSKCRKTFKRWKSFLNHQQTHSREK 352
Score = 29.3 bits (64), Expect = 8.5
Identities = 10/26 (38%), Positives = 13/26 (49%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRE 78
+ C C++ F AL HQ HK E
Sbjct: 1044 YKCGLCERSFVEKSALSRHQRVHKNE 1069
Score = 29.3 bits (64), Expect = 8.5
Identities = 12/27 (44%), Positives = 14/27 (51%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRER 79
+ C C KKF+ AL HQ H ER
Sbjct: 164 YQCVECQKKFTERSALVNHQRTHTGER 190
>Z473_HUMAN (Q8WTR7) Zinc finger protein 473 (Zinc finger protein
100 homolog) (Zfp-100)
Length = 871
Score = 35.8 bits (81), Expect = 0.091
Identities = 16/32 (50%), Positives = 19/32 (59%)
Query: 46 PEQKVVEFSCNYCDKKFSTSQALGGHQNAHKR 77
PEQK F CN C+K FS S+ L H+ H R
Sbjct: 555 PEQKEKCFKCNKCEKTFSCSKYLTQHERIHTR 586
Score = 33.1 bits (74), Expect = 0.59
Identities = 20/78 (25%), Positives = 34/78 (42%)
Query: 2 PPTSESHVEDFLTLRLGTHTKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKK 61
P ++ S ED + + T + +K E G S+ + + ++ + C+ C K
Sbjct: 158 PVSTVSTGEDSMVHNVSEKTLTPAKSKEYRGEFFSYSDHSQQDSVQEGEKPYQCSECGKS 217
Query: 62 FSTSQALGGHQNAHKRER 79
FS S L H H RE+
Sbjct: 218 FSGSYRLTQHWITHTREK 235
Score = 32.3 bits (72), Expect = 1.0
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 2/71 (2%)
Query: 40 TLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRF 99
T E + + V F C+ C K F S L HQ H R R+ K E + L ++
Sbjct: 578 TQHERIHTRGVKPFECDQCGKAFGQSTRLIHHQRIHSRVRLYKWGEQGKAISSASL-IKL 636
Query: 100 SSFIHYQGHSY 110
SF H + H +
Sbjct: 637 QSF-HTKEHPF 646
Score = 29.3 bits (64), Expect = 8.5
Identities = 14/30 (46%), Positives = 14/30 (46%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLK 82
F CN C K FS S L HQ H E K
Sbjct: 646 FKCNECGKTFSHSAHLSKHQLIHAGENPFK 675
>FZF1_YEAST (P32805) Zinc finger protein FZF1 (Sulphite resistance
protein 1)
Length = 299
Score = 35.8 bits (81), Expect = 0.091
Identities = 15/37 (40%), Positives = 22/37 (58%)
Query: 54 SCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRRE 90
+C C K+F T+Q L H N+H+R+ L DR+ E
Sbjct: 73 ACTLCQKRFVTNQQLRRHLNSHERKSKLASRIDRKHE 109
>ZG66_XENLA (P18733) Gastrula zinc finger protein XLCGF66.1
(Fragment)
Length = 606
Score = 35.4 bits (80), Expect = 0.12
Identities = 27/67 (40%), Positives = 32/67 (47%), Gaps = 8/67 (11%)
Query: 21 TKST--SKPLENGGHS----SSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNA 74
TKST SK N GH S N P QK FSC+ C K FS +L HQ
Sbjct: 237 TKSTLHSKDSCNEGHKHLSHKSDYNKHQNPHKRQK--SFSCSKCGKCFSNLTSLHCHQKT 294
Query: 75 HKRERVL 81
HK +++L
Sbjct: 295 HKGKKLL 301
>ZF90_MOUSE (Q61967) Zinc finger protein 90 (Zfp-90) (Zinc finger
protein NK10)
Length = 636
Score = 35.4 bits (80), Expect = 0.12
Identities = 14/27 (51%), Positives = 17/27 (62%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRER 79
F CN C K F S +LG H+NAH E+
Sbjct: 278 FECNVCGKAFRHSSSLGQHENAHTGEK 304
>ZF90_HUMAN (Q8TF47) Zinc finger protein 90 homolog (Zfp-90)
(Fragment)
Length = 468
Score = 35.4 bits (80), Expect = 0.12
Identities = 14/27 (51%), Positives = 17/27 (62%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRER 79
F CN C K F S +LG H+NAH E+
Sbjct: 114 FECNVCGKAFRHSSSLGQHENAHTGEK 140
Score = 32.0 bits (71), Expect = 1.3
Identities = 13/28 (46%), Positives = 16/28 (56%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERV 80
+SC C K FS S AL HQ H R ++
Sbjct: 441 YSCKECGKNFSRSSALTKHQRIHTRNKL 468
>ZG48_XENLA (P18723) Gastrula zinc finger protein XLCGF48.2
(Fragment)
Length = 647
Score = 35.0 bits (79), Expect = 0.15
Identities = 24/80 (30%), Positives = 32/80 (40%), Gaps = 3/80 (3%)
Query: 3 PTSESHVEDF--LTLRLGTHTKSTSK-PLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCD 59
P E H D+ +T+ T TS ENG + SS + V F+CN C
Sbjct: 264 PYDEEHFSDYRIITVTDPTQVAGTSPHSTENGEYDSSMAACTSQLSKSALNVMFNCNVCH 323
Query: 60 KKFSTSQALGGHQNAHKRER 79
F+ L HQ H E+
Sbjct: 324 TAFNNKSNLVRHQRIHTGEK 343
>Z214_HUMAN (Q9UL59) Zinc finger protein 214 (BWSCR2 associated
zinc-finger protein 1) (BAZ 1)
Length = 606
Score = 35.0 bits (79), Expect = 0.15
Identities = 27/88 (30%), Positives = 34/88 (37%), Gaps = 23/88 (26%)
Query: 51 VEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQ---- 106
V +SCN C K FS +L HQ H E+ K E D L +S +H
Sbjct: 301 VPYSCNACGKSFSQISSLHNHQRVHTEEKFYK--------IECDKDLSRNSLLHIHQRLH 352
Query: 107 -----------GHSYFRSANLHNPIGVH 123
G S+ RS+ LH VH
Sbjct: 353 IGEKPFKCNQCGKSFNRSSVLHVHQRVH 380
Score = 32.7 bits (73), Expect = 0.77
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 1/58 (1%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSY 110
F CN C K F+ S L HQ H E+ K ++ + LR +H SY
Sbjct: 358 FKCNQCGKSFNRSSVLHVHQRVHTGEKPY-KCDECGKGFSQSSNLRIHQLVHTGEKSY 414
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,781,453
Number of Sequences: 164201
Number of extensions: 1327036
Number of successful extensions: 7024
Number of sequences better than 10.0: 243
Number of HSP's better than 10.0 without gapping: 190
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 5096
Number of HSP's gapped (non-prelim): 1913
length of query: 236
length of database: 59,974,054
effective HSP length: 107
effective length of query: 129
effective length of database: 42,404,547
effective search space: 5470186563
effective search space used: 5470186563
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.5 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146554.5


